Deep sequencing of the murine olfactory receptor neuron transcriptome
- PMID: 25590618
- PMCID: PMC4295871
- DOI: 10.1371/journal.pone.0113170
Deep sequencing of the murine olfactory receptor neuron transcriptome
Abstract
The ability of animals to sense and differentiate among thousands of odorants relies on a large set of olfactory receptors (OR) and a multitude of accessory proteins within the olfactory epithelium (OE). ORs and related signaling mechanisms have been the subject of intensive studies over the past years, but our knowledge regarding olfactory processing remains limited. The recent development of next generation sequencing (NGS) techniques encouraged us to assess the transcriptome of the murine OE. We analyzed RNA from OEs of female and male adult mice and from fluorescence-activated cell sorting (FACS)-sorted olfactory receptor neurons (ORNs) obtained from transgenic OMP-GFP mice. The Illumina RNA-Seq protocol was utilized to generate up to 86 million reads per transcriptome. In OE samples, nearly all OR and trace amine-associated receptor (TAAR) genes involved in the perception of volatile amines were detectably expressed. Other genes known to participate in olfactory signaling pathways were among the 200 genes with the highest expression levels in the OE. To identify OE-specific genes, we compared olfactory neuron expression profiles with RNA-Seq transcriptome data from different murine tissues. By analyzing different transcript classes, we detected the expression of non-olfactory GPCRs in ORNs and established an expression ranking for GPCRs detected in the OE. We also identified other previously undescribed membrane proteins as potential new players in olfaction. The quantitative and comprehensive transcriptome data provide a virtually complete catalogue of genes expressed in the OE and present a useful tool to uncover candidate genes involved in, for example, olfactory signaling, OR trafficking and recycling, and proliferation.
Conflict of interest statement
Figures






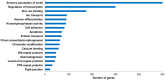

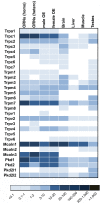
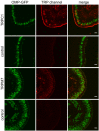
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

