Reactivation of epigenetically silenced miR-512 and miR-373 sensitizes lung cancer cells to cisplatin and restricts tumor growth
- PMID: 25591738
- PMCID: PMC4495356
- DOI: 10.1038/cdd.2014.221
Reactivation of epigenetically silenced miR-512 and miR-373 sensitizes lung cancer cells to cisplatin and restricts tumor growth
Abstract
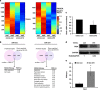
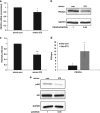
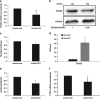
MicroRNAs (miRs) regulate a variety of cellular processes, and their impaired expression is involved in cancer. Silencing of tumor-suppressive miRs in cancer can occur through epigenetic modifications, including DNA methylation and histone deacetylation. We performed comparative miR profiling on cultured lung cancer cells before and after treatment with 5'aza-deoxycytidine plus Trichostatin A to reverse DNA methylation and histone deacetylation, respectively. Several tens of miRs were strongly induced by such 'epigenetic therapy'. Two representatives, miR-512-5p (miR-512) and miR-373, were selected for further analysis. Both miRs were secreted in exosomes. Re-expression of both miRs augmented cisplatin-induced apoptosis and inhibited cell migration; miR-512 also reduced cell proliferation. TEAD4 mRNA was confirmed as a direct target of miR-512; likewise, miR-373 was found to target RelA and PIK3CA mRNA directly. Our results imply that miR-512 and miR-373 exert cell-autonomous and non-autonomous tumor-suppressive effects in lung cancer cells, where their re-expression may benefit epigenetic cancer therapy.
Figures
Similar articles
-
MiR-152 and miR-185 co-contribute to ovarian cancer cells cisplatin sensitivity by targeting DNMT1 directly: a novel epigenetic therapy independent of decitabine.Oncogene. 2014 Jan 16;33(3):378-86. doi: 10.1038/onc.2012.575. Epub 2013 Jan 14. Oncogene. 2014. PMID: 23318422
-
Suberoylanilide hydroxamic acid inhibits growth of head and neck cancer cell lines by reactivation of tumor suppressor microRNAs.Oral Oncol. 2016 May;56:32-9. doi: 10.1016/j.oraloncology.2016.02.015. Epub 2016 Mar 19. Oral Oncol. 2016. PMID: 27086484 Free PMC article.
-
Genome-wide pharmacologic unmasking identifies tumor suppressive microRNAs in multiple myeloma.Oncotarget. 2015 Sep 22;6(28):26508-18. doi: 10.18632/oncotarget.4769. Oncotarget. 2015. PMID: 26164366 Free PMC article.
-
Epigenetic regulation of miR-129-2 and its effects on the proliferation and invasion in lung cancer cells.J Cell Mol Med. 2015 Sep;19(9):2172-80. doi: 10.1111/jcmm.12597. Epub 2015 Jun 17. J Cell Mol Med. 2015. PMID: 26081366 Free PMC article.
-
Epigenetic silencing of Aristaless-like homeobox-4, a potential tumor suppressor gene associated with lung cancer.Int J Cancer. 2014 Mar 15;134(6):1311-22. doi: 10.1002/ijc.28472. Epub 2013 Nov 11. Int J Cancer. 2014. PMID: 24037716
Cited by
-
The crosstalk between reactive oxygen species and noncoding RNAs: from cancer code to drug role.Mol Cancer. 2022 Jan 26;21(1):30. doi: 10.1186/s12943-021-01488-3. Mol Cancer. 2022. PMID: 35081965 Free PMC article. Review.
-
Roles and mechanisms of exosomal non-coding RNAs in human health and diseases.Signal Transduct Target Ther. 2021 Nov 10;6(1):383. doi: 10.1038/s41392-021-00779-x. Signal Transduct Target Ther. 2021. PMID: 34753929 Free PMC article. Review.
-
Response to neoadjuvant chemotherapy in breast cancer: do microRNAs matter?Discov Oncol. 2022 Jun 7;13(1):43. doi: 10.1007/s12672-022-00507-z. Discov Oncol. 2022. PMID: 35668332 Free PMC article.
-
Decitabine Treatment Induces a Viral Mimicry Response in Cervical Cancer Cells and Further Sensitizes Cells to Chemotherapy.Int J Mol Sci. 2022 Nov 14;23(22):14042. doi: 10.3390/ijms232214042. Int J Mol Sci. 2022. PMID: 36430521 Free PMC article.
-
Understanding the CREB1-miRNA feedback loop in human malignancies.Tumour Biol. 2016 Jul;37(7):8487-502. doi: 10.1007/s13277-016-5050-x. Epub 2016 Apr 9. Tumour Biol. 2016. PMID: 27059735 Review.
References
-
- Liu HP, Isaac WuHD, Chang JW, Wu YC, Yang HY, Chen YT, et al. Prognostic implications of epidermal growth factor receptor and KRAS gene mutations and epidermal growth factor receptor gene copy numbers in patients with surgically resectable non-small cell lung cancer in Taiwan. J Thorac Oncol. 2010;5:1175–1184. - PubMed
-
- Bergman Y, Cedar H. DNA methylation dynamics in health and disease. Nat Struct Mol Biol. 2013;20:274–281. - PubMed
-
- Aran D, Hellman A. DNA methylation of transcriptional enhancers and cancer predisposition. Cell. 2013;154:11–13. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

