Avoiding misannotation of in-source fragmentation products as cellular metabolites in liquid chromatography-mass spectrometry-based metabolomics
- PMID: 25591916
- PMCID: PMC4354698
- DOI: 10.1021/ac504118y
Avoiding misannotation of in-source fragmentation products as cellular metabolites in liquid chromatography-mass spectrometry-based metabolomics
Abstract
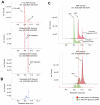
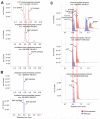
Liquid chromatography-mass spectrometry (LC-MS) technology allows for rapid quantitation of cellular metabolites, with metabolites identified by mass spectrometry and chromatographic retention time. Recently, with the development of rapid scanning high-resolution high accuracy mass spectrometers and the desire for high throughput screening, minimal or no chromatographic separation has become increasingly popular. When analyzing complex cellular extracts, however, the lack of chromatographic separation could potentially result in misannotation of structurally related metabolites. Here, we show that, even using electrospray ionization, a soft ionization method, in-source fragmentation generates unwanted byproducts of identical mass to common metabolites. For example, nucleotide-triphosphates generate nucleotide-diphosphates, and hexose-phosphates generate triose-phosphates. We evaluated yeast intracellular metabolite extracts and found more than 20 cases of in-source fragments that mimic common metabolites. Accordingly, chromatographic separation is required for accurate quantitation of many common cellular metabolites.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

