Analysis of the ORFK1 hypervariable regions reveal distinct HHV-8 clustering in Kaposi's sarcoma and non-Kaposi's cases
- PMID: 25592960
- PMCID: PMC4311464
- DOI: 10.1186/s13046-014-0119-0
Analysis of the ORFK1 hypervariable regions reveal distinct HHV-8 clustering in Kaposi's sarcoma and non-Kaposi's cases
Abstract
Background: Classical Kaposi's Sarcoma (cKS) is a rare vascular tumor, which develops in subjects infected with Human Herpesvirus-8 (HHV-8). Beside the host predisposing factors, viral genetic variants might possibly be related to disease development. The aim of this study was to identify HHV-8 variants in patients with cKS or in HHV-8 infected subjects either asymptomatic or with cKS-unrelated cutaneous lymphoproliferative disorders.
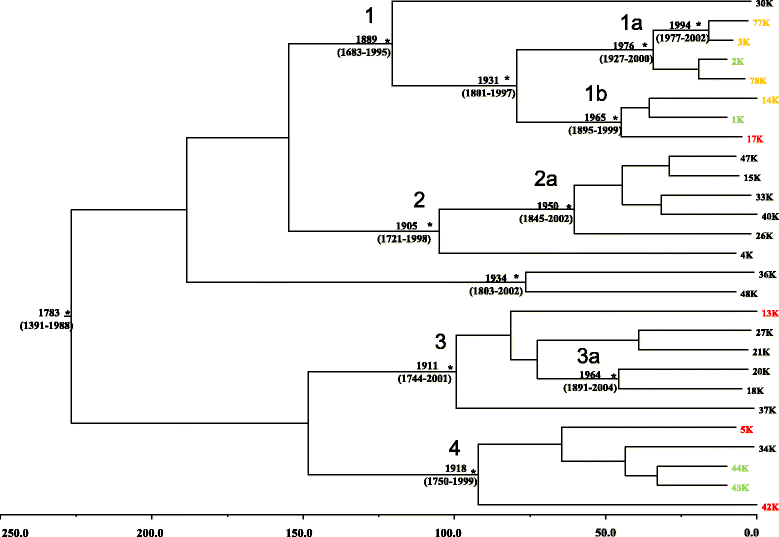
Methods: The VR1 and VR2 regions of the ORF K1 sequence were analyzed in samples (peripheral blood and/or lesional tissue) collected between 2000 and 2010 from 27 subjects with HHV-8 infection, established by the presence of anti-HHV-8 antibodies. On the basis of viral genotyping, a phylogenetic analysis and a time-scaled evaluation were performed.
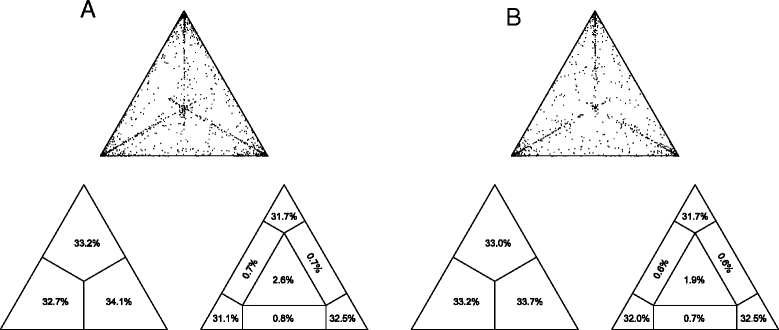
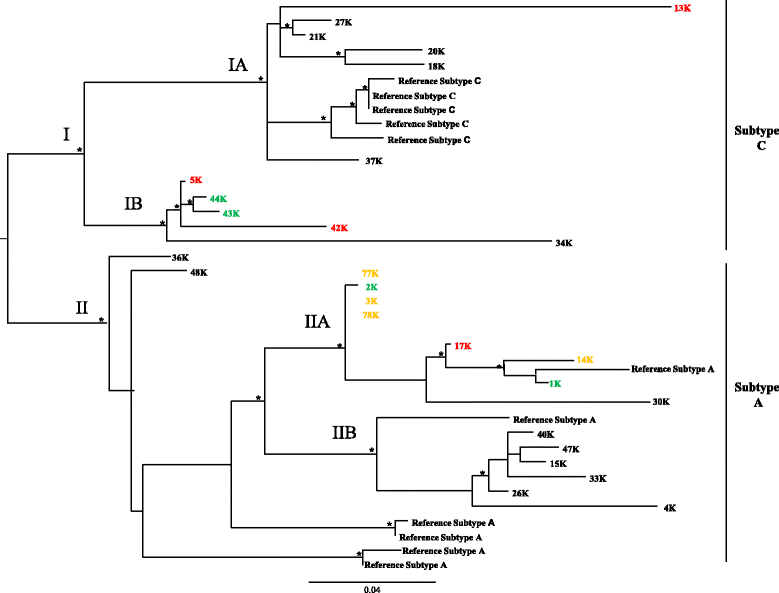
Results: Two main clades of HHV-8, corresponding to A and C subtypes, were identified. Moreover, for each subtype, two main clusters were found distinctively associated to cKS or non-cKS subjects. Selective pressure analysis showed twelve sites of the K1 coding gene (VR1 and VR2 regions) under positive selective pressure and one site under negative pressure.
Conclusion: Thus, present data suggest that HHV-8 genetic variants may influence the susceptibility to cKS in individuals with HHV-8 infection.
Figures
References
-
- Tamburro KM, Yang D, Poisson J, Fedoriw Y, Roy D, Lucas A, et al. Vironome of kaposi sarcoma associated herpesvirus-inflammatory cytokine syndrome in an AIDS patient reveals co-infection of human herpesvirus 8 and human herpesvirus 6A. Virology. 2012;433:220–5. doi: 10.1016/j.virol.2012.08.014. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

