The neXtProt knowledgebase on human proteins: current status
- PMID: 25593349
- PMCID: PMC4383972
- DOI: 10.1093/nar/gku1178
The neXtProt knowledgebase on human proteins: current status
Abstract
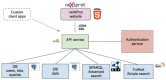
neXtProt (http://www.nextprot.org) is a human protein-centric knowledgebase developed at the SIB Swiss Institute of Bioinformatics. Focused solely on human proteins, neXtProt aims to provide a state of the art resource for the representation of human biology by capturing a wide range of data, precise annotations, fully traceable data provenance and a web interface which enables researchers to find and view information in a comprehensive manner. Since the introductory neXtProt publication, significant advances have been made on three main aspects: the representation of proteomics data, an extended representation of human variants and the development of an advanced search capability built around semantic technologies. These changes are presented in the current neXtProt update.
© The Author(s) 2014. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
-
- Bastian F., Parmentier G., Roux J., Moretti S., Laudet V., Robinson-Rechavi M. Data Integration in the Life Sciences. Vol. 5109. Springer Berlin/Heidelberg: 2008. pp. 124–131.
-
- Uhlen M., Oksvold P., Fagerberg L., Lundberg E., Jonasson K., Forsberg M., Zwahlen M., Kampf C., Wester K., Hober S., et al. Towards a knowledge-based Human Protein Atlas. Nat. Biotechnol. 2010;28:1248–1250. - PubMed
-
- Farrah T., Deutsch E.W., Omenn G.S., Sun Z., Watts J.D., Yamamoto T., Shteynberg D., Harris M.M., Moritz R.L. State of the human proteome in 2013 as viewed through PeptideAtlas: comparing the kidney, urine, and plasma proteomes for the biology- and disease-driven Human Proteome Project. J. Proteome Res. 2014;3:60–75. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

