Luciferase fragment complementation imaging in preclinical cancer studies
- PMID: 25594026
- PMCID: PMC4278313
- DOI: 10.18632/oncoscience.45
Luciferase fragment complementation imaging in preclinical cancer studies
Abstract
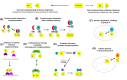
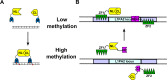
The luciferase fragment complementation assay (LFCA) enables molecular events to be non-invasively imaged in live cells in vitro and in vivo in a comparatively cheap and safe manner. It is a development of previous enzyme complementation assays in which reporter genes are split into two, individually enzymatically inactive, fragments that are able to complement one another upon interaction. This complementation can be used to externally visualize cellular activities. In recent years, the number of studies which have used LFCAs to probe questions relevant to cancer have increased, and this review summarizes the most significant and interesting of these. In particular, it focuses on work conducted on the epidermal growth factor, nuclear and chemokine receptor families, and intracellular signaling pathways, including IP3, cAMP, Akt, cMyc, NRF2 and Rho GTPases. LFCAs which have been developed to image DNA methylation and detect RNA transcripts are also discussed.
Keywords: Cancer; Imaging; Luciferase.
Figures



References
-
- Hastings JW. Chemistries and colors of bioluminescent reactions: a review. Gene. 1996;173:5–11. - PubMed
-
- Hastings JW. Biological diversity, chemical mechanisms, and the evolutionary origins of bioluminescent systems. Journal of Molecular Evolution. 1983;19(5):309–321. - PubMed
-
- Zhao H, Doyle TC, Coquoz O, Kalish F, Rice BW, Contag CH. Emission spectra of bioluminescent reporters and interaction with mammalian tissue determine the sensitivity of detection in vivo. J Biomed Opt. 2005;10(4):41210. - PubMed
-
- Weissleder R, Ntziachristos V. Shedding light onto live molecular targets. Nat Med. 2003;9(1):123–128. - PubMed
-
- Tannous BA, Kim DE, Fernandez JL, Weissleder R, Breakefield XO. Codon-optimized Gaussia luciferase cDNA for mammalian gene expression in culture and in vivo. Molecular therapy: the journal of the American Society of Gene Therapy. 2005;11(3):435–443. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

