Conformational changes of elongation factor G on the ribosome during tRNA translocation
- PMID: 25594181
- PMCID: PMC4297320
- DOI: 10.1016/j.cell.2014.11.049
Conformational changes of elongation factor G on the ribosome during tRNA translocation
Abstract
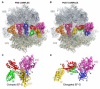
The universally conserved GTPase elongation factor G (EF-G) catalyzes the translocation of tRNA and mRNA on the ribosome after peptide bond formation. Despite numerous studies suggesting that EF-G undergoes extensive conformational rearrangements during translocation, high-resolution structures exist for essentially only one conformation of EF-G in complex with the ribosome. Here, we report four atomic-resolution crystal structures of EF-G bound to the ribosome programmed in the pre- and posttranslocational states and to the ribosome trapped by the antibiotic dityromycin. We observe a previously unseen conformation of EF-G in the pretranslocation complex, which is independently captured by dityromycin on the ribosome. Our structures provide insights into the conformational space that EF-G samples on the ribosome and reveal that tRNA translocation on the ribosome is facilitated by a structural transition of EF-G from a compact to an elongated conformation, which can be prevented by the antibiotic dityromycin.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures



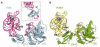
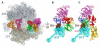
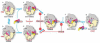
References
-
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. - PubMed
-
- Agrawal RK, Heagle AB, Penczek P, Grassucci RA, Frank J. EF-G-dependent GTP hydrolysis induces translocation accompanied by large conformational changes in the 70S ribosome. Nat Struct Biol. 1999;6:643–647. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

