Monitoring interactions and dynamics of endogenous beta-catenin with intracellular nanobodies in living cells
- PMID: 25595278
- PMCID: PMC4349989
- DOI: 10.1074/mcp.M114.044016
Monitoring interactions and dynamics of endogenous beta-catenin with intracellular nanobodies in living cells
Abstract
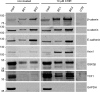
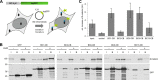
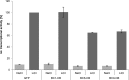
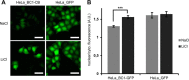
β-catenin is the key component of the canonical Wnt pathway and plays a crucial role in a multitude of developmental and homeostatic processes. The different tasks of β-catenin are orchestrated by its subcellular localization and participation in multiprotein complexes. To gain a better understanding of β-catenin's role in living cells we have generated a new set of single domain antibodies, referred to as nanobodies, derived from heavy chain antibodies of camelids. We selected nanobodies recognizing the N-terminal, core or C-terminal domain of β-catenin and applied these new high-affinity binders as capture molecules in sandwich immunoassays and co-immunoprecipitations of endogenous β-catenin complexes. In addition, we engineered intracellularly functional anti-β-catenin chromobodies by combining the binding moieties of the nanobodies with fluorescent proteins. For the first time, we were able to visualize the subcellular localization and nuclear translocation of endogenous β-catenin in living cells using these chromobodies. Moreover, the chromobody signal allowed us to trace the accumulation of diffusible, hypo-phosphorylated β-catenin in response to compound treatment in real time using High Content Imaging. The anti-β-catenin nanobodies and chromobodies characterized in this study are versatile tools that enable a novel and unique approach to monitor the dynamics of subcellular β-catenin in biochemical and cell biological assays.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures


References
-
- Takeichi M. (1995) Morphogenetic roles of classic cadherins. Curr. Opin. Cell Biol. 7, 619–627 - PubMed
-
- Korinek V., Barker N., Morin P. J., van Wichen D., de Weger R., Kinzler K. W., Vogelstein B., Clevers H. (1997) Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC−/− colon carcinoma. Science 275, 1784–1787 - PubMed
-
- Behrens J., Lustig B. (2004) The Wnt connection to tumorigenesis. Int. J. Dev. Biol. 48, 477–487 - PubMed
-
- Hart M., Concordet J. P., Lassot I., Albert I., del los Santos R., Durand H., Perret C., Rubinfeld B., Margottin F., Benarous R., Polakis P. (1999) The F-box protein beta-TrCP associates with phosphorylated beta-catenin and regulates its activity in the cell. Curr. Biol. 9, 207–210 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

