Quantifying on- and off-target genome editing
- PMID: 25595557
- PMCID: PMC4308725
- DOI: 10.1016/j.tibtech.2014.12.001
Quantifying on- and off-target genome editing
Abstract
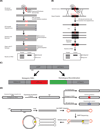
Genome editing with engineered nucleases is a rapidly growing field thanks to transformative technologies that allow researchers to precisely alter genomes for numerous applications including basic research, biotechnology, and human gene therapy. While the ability to make precise and controlled changes at specified sites throughout the genome has grown tremendously in recent years, we still lack a comprehensive and standardized battery of assays for measuring the different genome editing outcomes created at endogenous genomic loci. Here we review the existing assays for quantifying on- and off-target genome editing and describe their utility in advancing the technology. We also highlight unmet assay needs for quantifying on- and off-target genome editing outcomes and discuss their importance for the genome editing field.
Keywords: CRISPR/Cas9; RNA-guided endonucleases; TALENs; ZFNs; gene editing; gene targeting; homologous recombination; nonhomologous end-joining.
Copyright © 2014 Elsevier Ltd. All rights reserved.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

