LCK over-expression drives STAT5 oncogenic signaling in PAX5 translocated BCP-ALL patients
- PMID: 25595912
- PMCID: PMC4359315
- DOI: 10.18632/oncotarget.2807
LCK over-expression drives STAT5 oncogenic signaling in PAX5 translocated BCP-ALL patients
Abstract
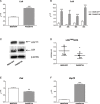
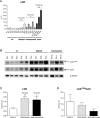
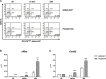
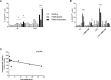
The PAX5 gene is altered in 30% of BCP-ALL patients and PAX5 chromosomal translocations account for 2-3% of cases. Although PAX5 fusion genes significantly affect the transcription of PAX5 target genes, their role in sustaining leukemia cell survival is poorly understood. In an in vitro model of PAX5/ETV6 leukemia, we demonstrated that Lck hyper-activation, and down-regulation of its negative regulator Csk, lead to STAT5 hyper-activation and consequently to the up-regulation of the downstream effectors, cMyc and Ccnd2. More important, cells from PAX5 translocated patients show LCK up-regulation and over-activation, as well as STAT5 hyper-phosphorylation, compared to PAX5 wt and PAX5 deleted cases. As in BCR/ABL1 positive ALL, the hyper-activation of STAT5 pathway can represent a survival signal in PAX5 translocated cells, alternative to the pre-BCR, which is down-regulated. The LCK inhibitor BIBF1120 selectively reverts this phenomenon both in the murine model and in leukemic primary cells. LCK inhibitor could therefore represent a suitable candidate drug to target this subgroup of ALL patients.
Figures

Similar articles
-
Molecular role of the PAX5-ETV6 oncoprotein in promoting B-cell acute lymphoblastic leukemia.EMBO J. 2017 Mar 15;36(6):718-735. doi: 10.15252/embj.201695495. Epub 2017 Feb 20. EMBO J. 2017. PMID: 28219927 Free PMC article.
-
PAX5 fusion genes in acute lymphoblastic leukemia: A literature review.Medicine (Baltimore). 2023 May 19;102(20):e33836. doi: 10.1097/MD.0000000000033836. Medicine (Baltimore). 2023. PMID: 37335685 Free PMC article. Review.
-
The correlation between Pax5 deletion and patients survival in Iranian children with precursor B-cell acute lymphocytic leukemia.Cell Mol Biol (Noisy-le-grand). 2017 Aug 30;63(8):19-22. doi: 10.14715/cmb/2017.63.8.4. Cell Mol Biol (Noisy-le-grand). 2017. PMID: 28886309
-
PAX5 fusion genes are frequent in poor risk childhood acute lymphoblastic leukaemia and can be targeted with BIBF1120.EBioMedicine. 2022 Sep;83:104224. doi: 10.1016/j.ebiom.2022.104224. Epub 2022 Aug 16. EBioMedicine. 2022. PMID: 35985167 Free PMC article.
-
GEMMs addressing Pax5 loss-of-function in childhood pB-ALL.Eur J Med Genet. 2016 Mar;59(3):166-72. doi: 10.1016/j.ejmg.2015.11.009. Epub 2015 Nov 26. Eur J Med Genet. 2016. PMID: 26626503 Review.
Cited by
-
Molecular role of the PAX5-ETV6 oncoprotein in promoting B-cell acute lymphoblastic leukemia.EMBO J. 2017 Mar 15;36(6):718-735. doi: 10.15252/embj.201695495. Epub 2017 Feb 20. EMBO J. 2017. PMID: 28219927 Free PMC article.
-
A Self-Activating IL-15 Chimeric Cytokine Receptor to Empower Cancer Immunotherapy.Immunotargets Ther. 2024 Oct 10;13:513-524. doi: 10.2147/ITT.S490498. eCollection 2024. Immunotargets Ther. 2024. PMID: 39403195 Free PMC article.
-
The Src family kinase LCK cooperates with oncogenic FLT3/ITD in cellular transformation.Sci Rep. 2017 Oct 23;7(1):13734. doi: 10.1038/s41598-017-14033-4. Sci Rep. 2017. PMID: 29062038 Free PMC article.
-
PAX5 fusion genes in acute lymphoblastic leukemia: A literature review.Medicine (Baltimore). 2023 May 19;102(20):e33836. doi: 10.1097/MD.0000000000033836. Medicine (Baltimore). 2023. PMID: 37335685 Free PMC article. Review.
-
New horizons in drug discovery of lymphocyte-specific protein tyrosine kinase (Lck) inhibitors: a decade review (2011-2021) focussing on structure-activity relationship (SAR) and docking insights.J Enzyme Inhib Med Chem. 2021 Dec;36(1):1574-1602. doi: 10.1080/14756366.2021.1937143. J Enzyme Inhib Med Chem. 2021. PMID: 34233563 Free PMC article. Review.
References
-
- Cobaleda C, Schebesta A, Delogu A, Busslinger M. Pax5: the guardian of B cell identity and function. Nature Immunology. 2007;8:463–470. - PubMed
-
- Mullighan CG, Goorha S, Radtke I, Miller CB, Coustan-Smith E, Dalton JD, Girtman K, Mathew S, Ma J, Pounds SB, Su X, Pui CH, Relling MV, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446:758–764. - PubMed
-
- Pui CH, Relling MV, Downing JR. Acute lymphoblastic leukemia. The New England journal of medicine. 2004;350:1535–1548. - PubMed
-
- Nebral K, Denk D, Attarbaschi A, Konig M, Mann G, Haas OA, Strehl S. Incidence and diversity of PAX5 fusion genes in childhood acute lymphoblastic leukemia. Leukemia. 2009;23:134–143. - PubMed
-
- Coyaud E, Struski S, Prade N, Familiades J, Eichner R, Quelen C, Bousquet M, Mugneret F, Talmant P, Pages MP, Lefebvre C, Penther D, Lippert E, et al. Wide diversity of PAX5 alterations in B-ALL: a Groupe Francophone de Cytogenetique Hematologique study. Blood. 2010;115:3089–3097. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

