Genetic architecture of artemisinin-resistant Plasmodium falciparum
- PMID: 25599401
- PMCID: PMC4545236
- DOI: 10.1038/ng.3189
Genetic architecture of artemisinin-resistant Plasmodium falciparum
Abstract
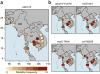
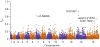
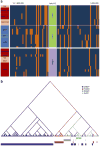
We report a large multicenter genome-wide association study of Plasmodium falciparum resistance to artemisinin, the frontline antimalarial drug. Across 15 locations in Southeast Asia, we identified at least 20 mutations in kelch13 (PF3D7_1343700) affecting the encoded propeller and BTB/POZ domains, which were associated with a slow parasite clearance rate after treatment with artemisinin derivatives. Nonsynonymous polymorphisms in fd (ferredoxin), arps10 (apicoplast ribosomal protein S10), mdr2 (multidrug resistance protein 2) and crt (chloroquine resistance transporter) also showed strong associations with artemisinin resistance. Analysis of the fine structure of the parasite population showed that the fd, arps10, mdr2 and crt polymorphisms are markers of a genetic background on which kelch13 mutations are particularly likely to arise and that they correlate with the contemporary geographical boundaries and population frequencies of artemisinin resistance. These findings indicate that the risk of new resistance-causing mutations emerging is determined by specific predisposing genetic factors in the underlying parasite population.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI101713/AI/NIAID NIH HHS/United States
- G0600718/MRC_/Medical Research Council/United Kingdom
- HHMI/Howard Hughes Medical Institute/United States
- 089276/WT_/Wellcome Trust/United Kingdom
- 090770/Z/09/Z/WT_/Wellcome Trust/United Kingdom
- MR/M006212/1/MRC_/Medical Research Council/United Kingdom
- ImNIH/Intramural NIH HHS/United States
- 090770/WT_/Wellcome Trust/United Kingdom
- 090532/Z/09/Z/WT_/Wellcome Trust/United Kingdom
- 098051/WT_/Wellcome Trust/United Kingdom
- 001/WHO_/World Health Organization/International
- 093956/WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

