Functional normalization of 450k methylation array data improves replication in large cancer studies
- PMID: 25599564
- PMCID: PMC4283580
- DOI: 10.1186/s13059-014-0503-2
Functional normalization of 450k methylation array data improves replication in large cancer studies
Abstract
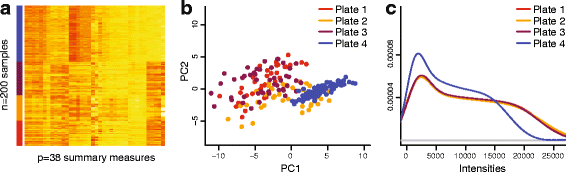
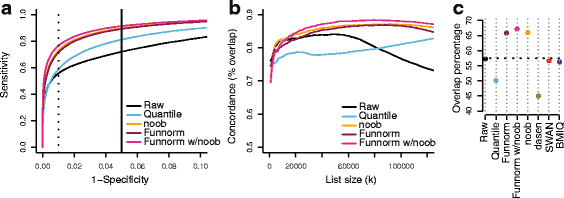
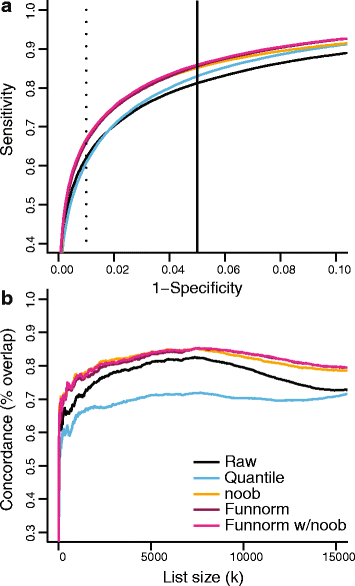
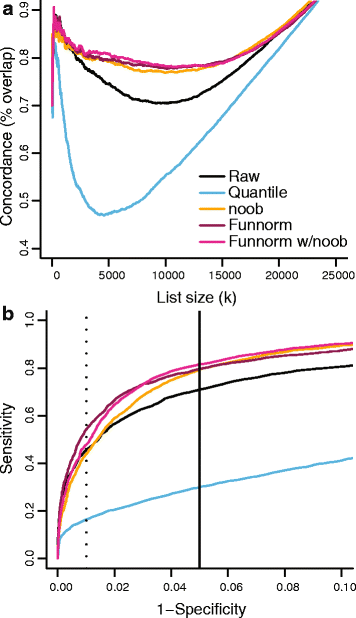
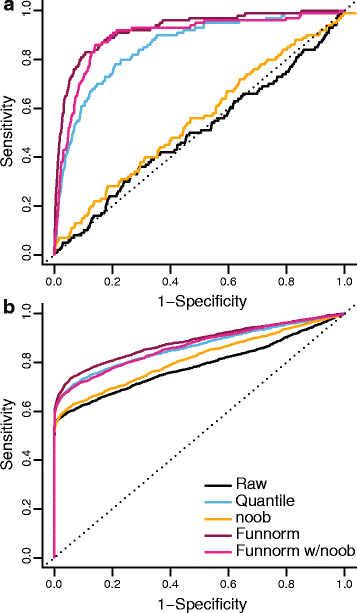
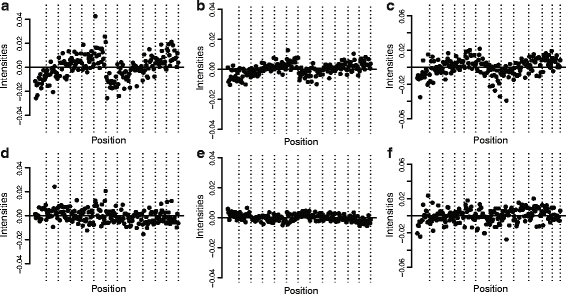
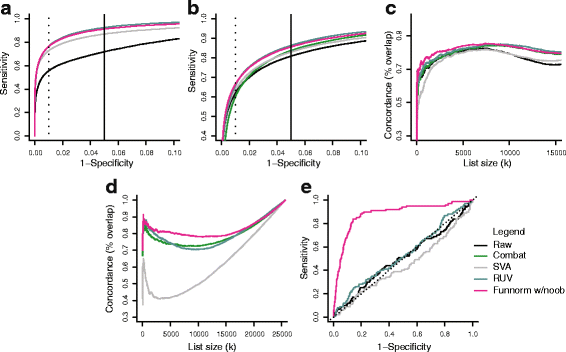
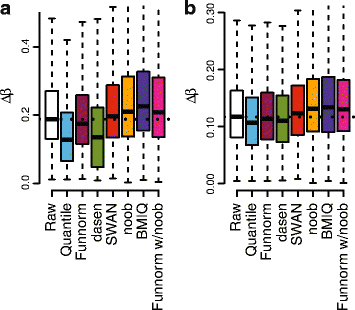
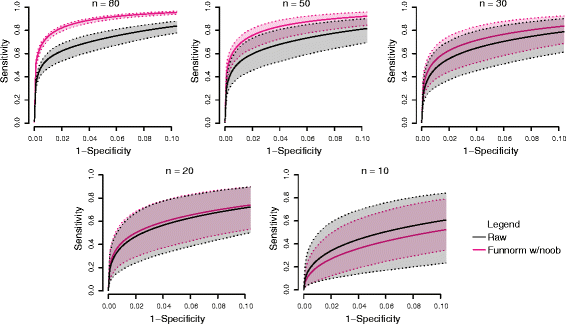
We propose an extension to quantile normalization that removes unwanted technical variation using control probes. We adapt our algorithm, functional normalization, to the Illumina 450k methylation array and address the open problem of normalizing methylation data with global epigenetic changes, such as human cancers. Using data sets from The Cancer Genome Atlas and a large case-control study, we show that our algorithm outperforms all existing normalization methods with respect to replication of results between experiments, and yields robust results even in the presence of batch effects. Functional normalization can be applied to any microarray platform, provided suitable control probes are available.
Figures

References
-
- Liu Y, Aryee MJ, Padyukov L, Fallin MD, Hesselberg E, Runarsson A, Reinius L, Acevedo N, Taub M, Ronninger M, Shchetynsky K, Scheynius A, Kere J, Alfredsson L, Klareskog L, Ekström TJ, Feinberg AP. Epigenome-wide association data implicate DNA methylation as an intermediary of genetic risk in rheumatoid arthritis. Nat Biotechnol. 2013;31:142–147. doi: 10.1038/nbt.2487. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

