Quantifying heterogeneity and dynamics of clonal fitness in response to perturbation
- PMID: 25600161
- PMCID: PMC5580929
- DOI: 10.1002/jcp.24888
Quantifying heterogeneity and dynamics of clonal fitness in response to perturbation
Abstract
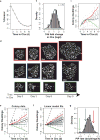
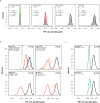
The dynamics of heterogeneous clonal lineages within a cell population, in aggregate, shape both normal and pathological biological processes. Studies of clonality typically relate the fitness of clones to their relative abundance, thus requiring long-term experiments and limiting conclusions about the heterogeneity of clonal fitness in response to perturbation. We present, for the first time, a method that enables a dynamic, global picture of clonal fitness within a mammalian cell population. This novel assay allows facile comparison of the structure of clonal fitness in a cell population across many perturbations. By utilizing high-throughput imaging, our methodology provides ample statistical power to define clonal fitness dynamically and to visualize the structure of perturbation-induced clonal fitness within a cell population. We envision that this technique will be a powerful tool to investigate heterogeneity in biological processes involving cell proliferation, including development and drug response.
© 2015 Wiley Periodicals, Inc.
Figures



References
-
- Anderson AR, Weaver AM, Cummings PT, Quaranta V. Tumor Morphology and Phenotypic Evolution Driven by Selective Pressure from the Microenvironment. Cell. 2006;127:905–915. - PubMed
-
- Azzalini A. The Skew-normal Distribution and Related Multivariate Families*. Scand J Stat. 2005;32:159–188.
-
- Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. Bacterial persistence as a phenotypic switch. Science. 2004;305:1622–1625. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

