The NAD(+)-dependent SIRT1 deacetylase translates a metabolic switch into regulatory epigenetics in skeletal muscle stem cells
- PMID: 25600643
- PMCID: PMC4320668
- DOI: 10.1016/j.stem.2014.12.004
The NAD(+)-dependent SIRT1 deacetylase translates a metabolic switch into regulatory epigenetics in skeletal muscle stem cells
Abstract
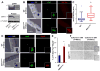
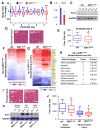
Stem cells undergo a shift in metabolic substrate utilization during specification and/or differentiation, a process that has been termed metabolic reprogramming. Here, we report that during the transition from quiescence to proliferation, skeletal muscle stem cells experience a metabolic switch from fatty acid oxidation to glycolysis. This reprogramming of cellular metabolism decreases intracellular NAD(+) levels and the activity of the histone deacetylase SIRT1, leading to elevated H4K16 acetylation and activation of muscle gene transcription. Selective genetic ablation of the SIRT1 deacetylase domain in skeletal muscle results in increased H4K16 acetylation and deregulated activation of the myogenic program in SCs. Moreover, mice with muscle-specific inactivation of the SIRT1 deacetylase domain display reduced myofiber size, impaired muscle regeneration, and derepression of muscle developmental genes. Overall, these findings reveal how metabolic cues can be mechanistically translated into epigenetic modifications that regulate skeletal muscle stem cell biology.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures





Comment in
-
SIRT1 synchs satellite cell metabolism with stem cell fate.Cell Stem Cell. 2015 Feb 5;16(2):103-4. doi: 10.1016/j.stem.2015.01.006. Cell Stem Cell. 2015. PMID: 25658362
References
-
- Abe S, Hirose D, Kado S, Iwanuma O, Saka H, Yanagisawa N, Ide Y. Increased expression of decorin during the regeneration stage of mdx mouse. Anat Sci Int. 2009;84:305–311. - PubMed
-
- Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, Tran H, Ross SE, Mostoslavsky R, Cohen HY, et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. - PubMed
-
- Chang NC, Rudnicki MA. Satellite cells: the architects of skeletal muscle. Current Topics in Developmental Biology. 2014;107:161–181. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

