A dynamically coupled allosteric network underlies binding cooperativity in Src kinase
- PMID: 25600932
- PMCID: PMC4300553
- DOI: 10.1038/ncomms6939
A dynamically coupled allosteric network underlies binding cooperativity in Src kinase
Abstract
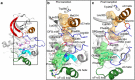
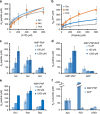
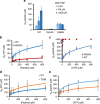
Protein tyrosine kinases are attractive drug targets because many human diseases are associated with the deregulation of kinase activity. However, how the catalytic kinase domain integrates different signals and switches from an active to an inactive conformation remains incompletely understood. Here we identify an allosteric network of dynamically coupled amino acids in Src kinase that connects regulatory sites to the ATP- and substrate-binding sites. Surprisingly, reactants (ATP and peptide substrates) bind with negative cooperativity to Src kinase while products (ADP and phosphopeptide) bind with positive cooperativity. We confirm the molecular details of the signal relay through the allosteric network by biochemical studies. Experiments on two additional protein tyrosine kinases indicate that the allosteric network may be largely conserved among these enzymes. Our work provides new insights into the regulation of protein tyrosine kinases and establishes a potential conduit by which resistance mutations to ATP-competitive kinase inhibitors can affect their activity.
Figures


References
-
- Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell 103, 211–225 (2000). - PubMed
-
- Manning G., Whyte D. B., Martinez R., Hunter T. & Sudarsanam S. The protein kinase complement of the human genome. Science 298, 1912–1934 (2002). - PubMed
-
- Hagerkvist R., Makeeva N., Elliman S. & Welsh N. Imatinib mesylate (Gleevec) protects against streptozotocin-induced diabetes and islet cell death in vitro. Cell Biol. Int. 30, 1013–1017 (2006). - PubMed
-
- Vlahovic G. & Crawford J. Activation of tyrosine kinases in cancer. Oncologist 8, 531–538 (2003). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

