Genome rearrangements can make and break small RNA genes
- PMID: 25601101
- PMCID: PMC4350180
- DOI: 10.1093/gbe/evv009
Genome rearrangements can make and break small RNA genes
Abstract
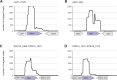
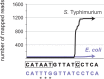
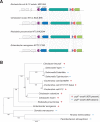
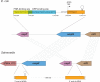
Small RNAs (sRNAs) are short, transcribed regulatory elements that are typically encoded in the intergenic regions (IGRs) of bacterial genomes. Several sRNAs, first recognized in Escherichia coli, are conserved among enteric bacteria, but because of the regulatory roles of sRNAs, differences in sRNA repertoires might be responsible for features that differentiate closely related species. We scanned the E. coli MG1655 and Salmonella enterica Typhimurium genomes for nonsyntenic IGRs as a potential source of uncharacterized, species-specific sRNAs and found that genome rearrangements have reconfigured several IGRs causing the disruption and formation of sRNAs. Within an IGR that is present in E. coli but was disrupted in Salmonella by a translocation event is an sRNA that is associated with the FNR/CRP global regulators and influences E. coli biofilm formation. A Salmonella-specific sRNA evolved de novo through point mutations that generated a σ(70) promoter sequence in an IGR that arose through genome rearrangement events. The differences in the sRNA pools among bacterial species have previously been ascribed to duplication, deletion, or horizontal acquisition. Here, we show that genomic rearrangements also contribute to this process by either disrupting sRNA-containing IGRs or creating IGRs in which novel sRNAs may evolve.
Keywords: E. coli; Salmonella; gene origination; intergenic regions; sRNA.
© The Author(s) 2015. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures


Similar articles
-
Emergence of New sRNAs in Enteric Bacteria is Associated with Low Expression and Rapid Evolution.J Mol Evol. 2017 Apr;84(4):204-213. doi: 10.1007/s00239-017-9793-9. Epub 2017 Apr 12. J Mol Evol. 2017. PMID: 28405712
-
Novel small RNA (sRNA) landscape of the starvation-stress response transcriptome of Salmonella enterica serovar typhimurium.RNA Biol. 2016;13(3):331-42. doi: 10.1080/15476286.2016.1144010. Epub 2016 Feb 6. RNA Biol. 2016. PMID: 26853797 Free PMC article.
-
Accessibility and conservation: general features of bacterial small RNA-mRNA interactions?RNA Biol. 2012 Jul;9(7):954-65. doi: 10.4161/rna.20294. Epub 2012 Jul 1. RNA Biol. 2012. PMID: 22767260 Free PMC article.
-
Trans-Acting Small RNAs and Their Effects on Gene Expression in Escherichia coli and Salmonella enterica.EcoSal Plus. 2020 Mar;9(1):10.1128/ecosalplus.ESP-0030-2019. doi: 10.1128/ecosalplus.ESP-0030-2019. EcoSal Plus. 2020. PMID: 32213244 Free PMC article. Review.
-
Origin, Evolution, and Loss of Bacterial Small RNAs.Microbiol Spectr. 2018 Apr;6(2):10.1128/microbiolspec.rwr-0004-2017. doi: 10.1128/microbiolspec.RWR-0004-2017. Microbiol Spectr. 2018. PMID: 29623872 Free PMC article. Review.
Cited by
-
Identification and Characterization of Novel Small RNAs in Rickettsia prowazekii.Front Microbiol. 2016 Jun 8;7:859. doi: 10.3389/fmicb.2016.00859. eCollection 2016. Front Microbiol. 2016. PMID: 27375581 Free PMC article.
-
A small RNA is functional in Escherichia fergusonii despite containing a large insertion.Microbiology (Reading). 2021 Oct;167(10):001099. doi: 10.1099/mic.0.001099. Microbiology (Reading). 2021. PMID: 34698627 Free PMC article.
-
Differential evolution in 3'UTRs leads to specific gene expression in Staphylococcus.Nucleic Acids Res. 2020 Mar 18;48(5):2544-2563. doi: 10.1093/nar/gkaa047. Nucleic Acids Res. 2020. PMID: 32016395 Free PMC article.
-
Functional annotation and distribution overview of RNA families in 27 Streptococcus agalactiae genomes.BMC Genomics. 2018 Jul 28;19(1):556. doi: 10.1186/s12864-018-4951-z. BMC Genomics. 2018. PMID: 30055586 Free PMC article.
-
RNA-Sequencing Analyses of Small Bacterial RNAs and their Emergence as Virulence Factors in Host-Pathogen Interactions.Int J Mol Sci. 2020 Feb 27;21(5):1627. doi: 10.3390/ijms21051627. Int J Mol Sci. 2020. PMID: 32120885 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

