Simultaneous impairment of mitochondrial fission and fusion reduces mitophagy and shortens replicative lifespan
- PMID: 25601284
- PMCID: PMC4298727
- DOI: 10.1038/srep07885
Simultaneous impairment of mitochondrial fission and fusion reduces mitophagy and shortens replicative lifespan
Abstract
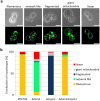
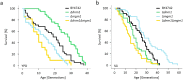
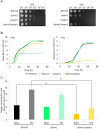
Aging of biological systems is accompanied by degeneration of mitochondrial functions. Different pathways are active to counteract the processes which lead to mitochondrial dysfunction. Mitochondrial dynamics, the fission and fusion of mitochondria, is one of these quality control pathways. Mitophagy, the controlled degradation of mitochondria, is another one. Here we show that these pathways are linked. A double deletion mutant of Saccharomyces cerevisiae in which two essential components of the fission and fusion machinery, Dnm1 and Mgm1, are simultaneously ablated, contain wild-type like filamentous mitochondria, but are characterized by impaired respiration, an increased sensitivity to different stressors, increased mitochondrial protein carbonylation, and a decrease in mitophagy and replicative lifespan. These data show that a balanced mitochondrial dynamics and not a filamentous mitochondrial morphotype per se is the key for a long lifespan and demonstrate a cross-talk between two different mitochondrial quality control pathways.
Figures



References
-
- Okamoto K. & Shaw J. M. Mitochondrial morphology and dynamics in yeast and multicellular eukaryotes. Annu. Rev. Genet. 39, 503–536 (2005). - PubMed
-
- Westermann B. Mitochondrial dynamics in model organisms: what yeasts, worms and flies have taught us about fusion and fission of mitochondria. Semin. Cell Dev. Biol. 21, 542–549 (2010). - PubMed
-
- Jones B. A. & Fangman W. L. Mitochondrial DNA maintenance in yeast requires a protein containing a region related to the GTP-binding domain of dynamin. Genes Dev. 6, 380–389 (1992). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

