Elimination of unfit cells maintains tissue health and prolongs lifespan
- PMID: 25601460
- PMCID: PMC4313366
- DOI: 10.1016/j.cell.2014.12.017
Elimination of unfit cells maintains tissue health and prolongs lifespan
Abstract
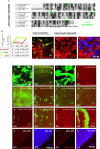
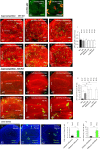
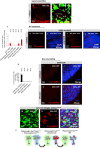
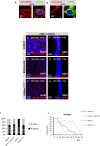
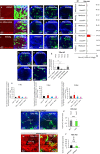
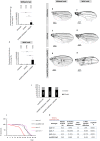
Viable yet damaged cells can accumulate during development and aging. Although eliminating those cells may benefit organ function, identification of this less fit cell population remains challenging. Previously, we identified a molecular mechanism, based on "fitness fingerprints" displayed on cell membranes, which allows direct fitness comparison among cells in Drosophila. Here, we study the physiological consequences of efficient cell selection for the whole organism. We find that fitness-based cell culling is naturally used to maintain tissue health, delay aging, and extend lifespan in Drosophila. We identify a gene, azot, which ensures the elimination of less fit cells. Lack of azot increases morphological malformations and susceptibility to random mutations and accelerates tissue degeneration. On the contrary, improving the efficiency of cell selection is beneficial for tissue health and extends lifespan.
Copyright © 2015 The Authors. Published by Elsevier Inc. All rights reserved.
Figures





References
-
- Baillon L., Basler K. Reflections on cell competition. Semin. Cell Dev. Biol. 2014;32:137–144. - PubMed
-
- Brodsky M.H., Nordstrom W., Tsang G., Kwan E., Rubin G.M., Abrams J.M. Drosophila p53 binds a damage response element at the reaper locus. Cell. 2000;101:103–113. - PubMed
-
- de la Cova C., Abril M., Bellosta P., Gallant P., Johnston L.A. Drosophila myc regulates organ size by inducing cell competition. Cell. 2004;117:107–116. - PubMed
-
- Fernández-Hernández I., Rhiner C., Moreno E. Adult neurogenesis in Drosophila. Cell Rep. 2013;3:1857–1865. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

