Alternative translational initiation of ATP sulfurylase underlying dual localization of sulfate assimilation pathways in plastids and cytosol in Arabidopsis thaliana
- PMID: 25601874
- PMCID: PMC4283515
- DOI: 10.3389/fpls.2014.00750
Alternative translational initiation of ATP sulfurylase underlying dual localization of sulfate assimilation pathways in plastids and cytosol in Arabidopsis thaliana
Abstract
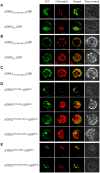
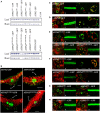
Plants assimilate inorganic sulfate into sulfur-containing vital metabolites. ATP sulfurylase (ATPS) is the enzyme catalyzing the key entry step of the sulfate assimilation pathway in both plastids and cytosol in plants. Arabidopsis thaliana has four ATPS genes (ATPS1, -2, -3, and -4) encoding ATPS pre-proteins containing N-terminal transit peptide sequences for plastid targeting, however, the genetic identity of the cytosolic ATPS has remained unverified. Here we show that Arabidopsis ATPS2 dually encodes plastidic and cytosolic ATPS isoforms, differentiating their subcellular localizations by initiating translation at AUG(Met1) to produce plastid-targeted ATPS2 pre-proteins or at AUG(Met52) or AUG(Met58) within the transit peptide to have ATPS2 stay in cytosol. Translational initiation of ATPS2 at AUG(Met52) or AUG(Met58) was verified by expressing a tandem-fused synthetic gene, ATPS2 (5'UTR-His12) :Renilla luciferase:ATPS2 (Ile13-Val77) :firefly luciferase, under a single constitutively active CaMV 35S promoter in Arabidopsis protoplasts and examining the activities of two different luciferases translated in-frame with split N-terminal portions of ATPS2. Introducing missense mutations at AUG(Met52) and AUG(Met58) significantly reduced the firefly luciferase activity, while AUG(Met52) was a relatively preferred site for the alternative translational initiation. The activity of luciferase fusion protein starting at AUG(Met52) or AUG(Met58) was not modulated by changes in sulfate conditions. The dual localizations of ATPS2 in plastids and cytosol were further evidenced by expression of ATPS2-GFP fusion proteins in Arabidopsis protoplasts and transgenic lines, while they were also under control of tissue-specific ATPS2 promoter activity found predominantly in leaf epidermal cells, guard cells, vascular tissues and roots.
Keywords: ATP sulfurylase; Arabidopsis; alternative translational initiation; dual localization; sulfur metabolism.
Figures


References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

