The deletion of rnhB in Mycobacterium smegmatis does not affect the level of RNase HII substrates or influence genome stability
- PMID: 25603150
- PMCID: PMC4300193
- DOI: 10.1371/journal.pone.0115521
The deletion of rnhB in Mycobacterium smegmatis does not affect the level of RNase HII substrates or influence genome stability
Abstract
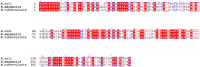
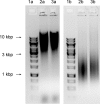
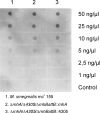
RNase HII removes RNA from RNA/DNA hybrids, such as single ribonucleotides and RNA primers generated during DNA synthesis. Both, RNase HII substrates and RNase HII deficiency have been associated with genome instability in several organisms, and genome instability is a major force leading to the acquisition of drug resistance in bacteria. Understanding the mechanisms that underlie this phenomenon is one of the challenges in identifying efficient methods to combat bacterial pathogens. The aim of the present study was set to investigate the role of rnhB, presumably encoding RNase HII, in maintaining genome stability in the M. tuberculosis model organism Mycobacterium smegmatis. We performed gene replacement through homologous recombination to obtain mutant strains of Mycobacterium smegmatis lacking the rnhB gene. The mutants did not present an altered phenotype, according to the growth rate in liquid culture or susceptibility to hydroxyurea, and did not show an increase in the spontaneous mutation rate, determined using the Luria-Delbrück fluctuation test for streptomycin resistance in bacteria. The mutants also did not present an increase in the level of RNase HII substrates, measured as the level of alkaline degradation of chromosomal DNA or determined through immunodetection. We conclude that proteins other than RnhB proteins efficiently remove RNase HII substrates in M. smegmatis. These results highlight differences in the basic biology between Mycobacteria and eukaryotes and between different species of bacteria.
Conflict of interest statement
Figures





Similar articles
-
Division of labor among Mycobacterium smegmatis RNase H enzymes: RNase H1 activity of RnhA or RnhC is essential for growth whereas RnhB and RnhA guard against killing by hydrogen peroxide in stationary phase.Nucleic Acids Res. 2017 Jan 9;45(1):1-14. doi: 10.1093/nar/gkw1046. Epub 2016 Nov 28. Nucleic Acids Res. 2017. PMID: 27899559 Free PMC article.
-
The C-Terminal Acid Phosphatase Module of the RNase HI Enzyme RnhC Controls Rifampin Sensitivity and Light-Dependent Colony Pigmentation of Mycobacterium smegmatis.J Bacteriol. 2023 Apr 25;205(4):e0043122. doi: 10.1128/jb.00431-22. Epub 2023 Mar 14. J Bacteriol. 2023. PMID: 36916909 Free PMC article.
-
RNase HI Is Essential for Survival of Mycobacterium smegmatis.PLoS One. 2015 May 12;10(5):e0126260. doi: 10.1371/journal.pone.0126260. eCollection 2015. PLoS One. 2015. PMID: 25965344 Free PMC article.
-
Synthetic lethal mutants in Escherichia coli define pathways necessary for survival with RNase H deficiency.J Bacteriol. 2023 Oct 26;205(10):e0028023. doi: 10.1128/jb.00280-23. Epub 2023 Oct 11. J Bacteriol. 2023. PMID: 37819120 Free PMC article.
-
Ribonuclease H: the enzymes in eukaryotes.FEBS J. 2009 Mar;276(6):1494-505. doi: 10.1111/j.1742-4658.2009.06908.x. Epub 2008 Feb 18. FEBS J. 2009. PMID: 19228196 Free PMC article. Review.
Cited by
-
DNA Replication in Mycobacterium tuberculosis.Microbiol Spectr. 2017 Mar;5(2):10.1128/microbiolspec.tbtb2-0027-2016. doi: 10.1128/microbiolspec.TBTB2-0027-2016. Microbiol Spectr. 2017. PMID: 28361736 Free PMC article. Review.
-
Division of labor among Mycobacterium smegmatis RNase H enzymes: RNase H1 activity of RnhA or RnhC is essential for growth whereas RnhB and RnhA guard against killing by hydrogen peroxide in stationary phase.Nucleic Acids Res. 2017 Jan 9;45(1):1-14. doi: 10.1093/nar/gkw1046. Epub 2016 Nov 28. Nucleic Acids Res. 2017. PMID: 27899559 Free PMC article.
-
Biochemical Characterization of Mycobacterium smegmatis RnhC (MSMEG_4305), a Bifunctional Enzyme Composed of Autonomous N-Terminal Type I RNase H and C-Terminal Acid Phosphatase Domains.J Bacteriol. 2015 Aug 1;197(15):2489-98. doi: 10.1128/JB.00268-15. Epub 2015 May 18. J Bacteriol. 2015. PMID: 25986906 Free PMC article.
-
The C-Terminal Acid Phosphatase Module of the RNase HI Enzyme RnhC Controls Rifampin Sensitivity and Light-Dependent Colony Pigmentation of Mycobacterium smegmatis.J Bacteriol. 2023 Apr 25;205(4):e0043122. doi: 10.1128/jb.00431-22. Epub 2023 Mar 14. J Bacteriol. 2023. PMID: 36916909 Free PMC article.
-
Large-Scale Photolithographic Synthesis of Chimeric DNA/RNA Hairpin Microarrays To Explore Sequence Specificity Landscapes of RNase HII Cleavage.Biochemistry. 2019 Nov 5;58(44):4389-4397. doi: 10.1021/acs.biochem.9b00806. Epub 2019 Oct 28. Biochemistry. 2019. PMID: 31631649 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

