Transcriptional regulation of bacterial virulence gene expression by molecular oxygen and nitric oxide
- PMID: 25603427
- PMCID: PMC4601167
- DOI: 10.4161/viru.27794
Transcriptional regulation of bacterial virulence gene expression by molecular oxygen and nitric oxide
Abstract
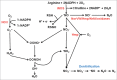
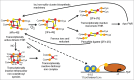
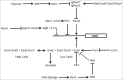
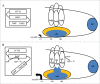
Molecular oxygen (O2) and nitric oxide (NO) are diatomic gases that play major roles in infection. The host innate immune system generates reactive oxygen species and NO as bacteriocidal agents and both require O2 for their production. Furthermore, the ability to adapt to changes in O2 availability is crucial for many bacterial pathogens, as many niches within a host are hypoxic. Pathogenic bacteria have evolved transcriptional regulatory systems that perceive these gases and respond by reprogramming gene expression. Direct sensors possess iron-containing co-factors (iron-sulfur clusters, mononuclear iron, heme) or reactive cysteine thiols that react with O2 and/or NO. Indirect sensors perceive the physiological effects of O2 starvation. Thus, O2 and NO act as environmental cues that trigger the coordinated expression of virulence genes and metabolic adaptations necessary for survival within a host. Here, the mechanisms of signal perception by key O2- and NO-responsive bacterial transcription factors and the effects on virulence gene expression are reviewed, followed by consideration of these aspects of gene regulation in two major pathogens, Staphylococcus aureus and Mycobacterium tuberculosis.
Keywords: AIP, autoinducer peptide; Arc, Aerobic respiratory control; FNR; FNR, fumarate nitrate reduction regulator; GAF, cGMP-specific phosphodiesterase-adenylyl cyclase-FhlA domain; Isc, iron–sulfur cluster biosynthesis machinery; Mycobacterium tuberculosis; NOX, NADPH oxidase; PAS, Per-Amt-Sim domain; RNS, reactive nitrogen species; ROS, reactive oxygen species; Staphylococcus aureus; TB, tuberculosis; WhiB-like proteins; iNOS, inducible nitric oxide synthase; iron–sulfur cluster; nitric oxide sensors; oxygen sensors.
Figures
References
-
- Imlay JA. Pathways of oxidative damage. Annu Rev Microbiol 2003; 57:395-418; PMID:14527285; http://dx.doi.org/10.1146/annurev.micro.57.030502.090938 - DOI - PubMed
-
- Imlay JA. Cellular defenses against superoxide and hydrogen peroxide. Annu Rev Biochem 2008; 77:755-76; PMID:18173371; http://dx.doi.org/10.1146/annurev.biochem.77.061606.161055 - DOI - PMC - PubMed
-
- Robinson MA, Baumgardner JE, Otto CM. Oxygen-dependent regulation of nitric oxide production by inducible nitric oxide synthase. Free Radic Biol Med 2011; 51:1952-65; PMID:21958548; http://dx.doi.org/10.1016/j.freeradbiomed.2011.08.034 - DOI - PubMed
-
- Bowman LA, McLean S, Poole RK, Fukuto JM. The diversity of microbial responses to nitric oxide and agents of nitrosative stress close cousins but not identical twins. Adv Microb Physiol 2011; 59:135-219; PMID:22114842; http://dx.doi.org/10.1016/B978-0-12-387661-4.00006-9 - DOI - PubMed
-
- Crack JC, Green J, Thomson AJ, Le Brun NE. Iron-sulfur cluster sensor-regulators. Curr Opin Chem Biol 2012; 16:35-44; PMID:22387135; http://dx.doi.org/10.1016/j.cbpa.2012.02.009 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
