Pyrosequencing analysis of a bacterial community associated with lava-formed soil from the Gotjawal forest in Jeju, Korea
- PMID: 25604185
- PMCID: PMC4398510
- DOI: 10.1002/mbo3.238
Pyrosequencing analysis of a bacterial community associated with lava-formed soil from the Gotjawal forest in Jeju, Korea
Abstract
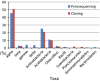
In this study, we analyzed the bacterial diversity in soils collected from Gyorae Gotjawal forest, where globally unique topography, geology, and ecological features support a forest grown on basalt flows from 110,000 to 120,000 years ago and 40,000 to 50,000 years ago. The soils at the site are fertile, with rocky areas, and are home to endangered species of plants and animals. Rainwater penetrates to the groundwater aquifer, which is composed of 34% organic matter containing rare types of soil and no soil profile. We determined the bacterial community composition using 116,475 reads from a 454-pyrosequencing analysis. This dataset included 12,621 operational taxonomic units at 3% dissimilarity, distributed among the following groups: Proteobacteria (56.2%) with 45.7% of α-Proteobacteria, Actinobacteria (25%), Acidobacteria (10.9%), Chloroflexi (2.4%), and Bacteroidetes (0.9%). In addition, 16S rRNA gene sequences were amplified using polymerase chain reaction and domain-specific primers to construct a clone library based on 142 bacterial clones. These clones were affiliated with the following groups: Proteobacteria (56%) with 51% of α-Proteobacteria, Acidobacteria (7.8%), Actinobacteria (17.6%), Chloroflexi (2.1%), Bacilli (1.4%), Cyanobacteria (2.8%), and Planctomycetes (1.4%). Within the phylum Proteobacteria, 56 of 80 clones were tentatively identified as 12 unclassified genera. Several new genera and a new family were discovered within the Actinobacteria clones. Results from 454-pyrosequencing revealed that 57% and 34% of the sequences belonged to undescribed genera and families, respectively. The characteristics of Gotjawal soil, which are determined by lava morphology, vegetation, and groundwater penetration, might be reflected in the bacterial community composition.
Keywords: 16S rRNA gene; Gotjawal forest soil; bacterial diversity; pyrosequencing.
© 2015 The Authors. MicrobiologyOpen published by John Wiley & Sons Ltd.
Figures



References
-
- Chan OC, Yang X, Fu Y, Feng Z, Sha L, Casper P, et al. 16S rRNA gene analyses of bacterial community structures in the soils of evergreen broad-leaved forests in south-west china. FEMS Microbiol. Ecol. 2006;58:247–259. - PubMed
-
- Crews TE, Kurina LM. Vitousek PM. Organic matter and nitrogen accumulation and nitrogen fixation during early ecosystem development in Hawaii. Biogeochemistry. 2001;52:259–279.
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

