Loss of osteoblast Runx3 produces severe congenital osteopenia
- PMID: 25605327
- PMCID: PMC4355527
- DOI: 10.1128/MCB.01106-14
Loss of osteoblast Runx3 produces severe congenital osteopenia
Abstract
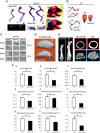
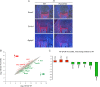
Congenital osteopenia is a bone demineralization condition that is associated with elevated fracture risk in human infants. Here we show that Runx3, like Runx2, is expressed in precommitted embryonic osteoblasts and that Runx3-deficient mice develop severe congenital osteopenia. Runx3-deficient osteoblast-specific (Runx3(fl/fl)/Col1α1-cre), but not chondrocyte-specific (Runx3(fl/fl)/Col1α2-cre), mice are osteopenic. This demonstrates that an osteoblastic cell-autonomous function of Runx3 is required for proper osteogenesis. Bone histomorphometry revealed that decreased osteoblast numbers and reduced mineral deposition capacity in Runx3-deficient mice cause this bone formation deficiency. Neonatal bone and cultured primary osteoblast analyses revealed a Runx3-deficiency-associated decrease in the number of active osteoblasts resulting from diminished proliferation and not from enhanced osteoblast apoptosis. These findings are supported by Runx3-null culture transcriptome analyses showing significant decreases in the levels of osteoblastic markers and increases in the levels of Notch signaling components. Thus, while Runx2 is mandatory for the osteoblastic lineage commitment, Runx3 is nonredundantly required for the proliferation of these precommitted cells, to generate adequate numbers of active osteoblasts. Human RUNX3 resides on chromosome 1p36, a region that is associated with osteoporosis. Therefore, RUNX3 might also be involved in human bone mineralization.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures




References
-
- Rauch F. 2005. Bone growth in length and width: the yin and yang of bone stability. J Musculoskelet Neuronal Interact 5:194–201. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
