The inhibitor protein (IF1) of the F1F0-ATPase modulates human osteosarcoma cell bioenergetics
- PMID: 25605724
- PMCID: PMC4358270
- DOI: 10.1074/jbc.M114.631788
The inhibitor protein (IF1) of the F1F0-ATPase modulates human osteosarcoma cell bioenergetics
Abstract
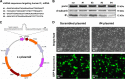
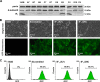
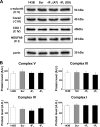
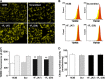
The bioenergetics of IF1 transiently silenced cancer cells has been extensively investigated, but the role of IF1 (the natural inhibitor protein of F1F0-ATPase) in cancer cell metabolism is still uncertain. To shed light on this issue, we established a method to prepare stably IF1-silenced human osteosarcoma clones and explored the bioenergetics of IF1 null cancer cells. We showed that IF1-silenced cells proliferate normally, consume glucose, and release lactate as controls do, and contain a normal steady-state ATP level. However, IF1-silenced cells displayed an enhanced steady-state mitochondrial membrane potential and consistently showed a reduced ADP-stimulated respiration rate. In the parental cells (i.e. control cells containing IF1) the inhibitor protein was found to be associated with the dimeric form of the ATP synthase complex, therefore we propose that the interaction of IF1 with the complex either directly, by increasing the catalytic activity of the enzyme, or indirectly, by improving the structure of mitochondrial cristae, can increase the oxidative phosphorylation rate in osteosarcoma cells grown under normoxic conditions.
Keywords: ATP Synthase; Bioenergetics; Cancer; F1F0-ATPase; IF1; Metabolism; Mitochondria; Oxidative Phosphorylation.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures





References
-
- Hanahan D., Weinberg R. A. (2011) Hallmarks of cancer: the next generation. Cell 144, 646–674 - PubMed
-
- Frezza C., Gottlieb E. (2009) Mitochondria in cancer: not just innocent bystanders. Semin. Cancer Biol. 19, 4–11 - PubMed
-
- Moreno-Sánchez R., Rodríguez-Enríquez S., Marín-Hernández A., Saavedra E. (2007) Energy metabolism in tumor cells. FEBS J. 274, 1393–1418 - PubMed
-
- Sánchez-Cenizo L., Formentini L., Aldea M., Ortega A. D., García-Huerta P., Sánchez-Aragó M., Cuezva J. M. (2010) Up-regulation of the ATPase inhibitory factor 1 (IF1) of the mitochondrial H+-ATP synthase in human tumors mediates the metabolic shift of cancer cells to a Warburg phenotype. J. Biol. Chem. 285, 25308–25313 - PMC - PubMed
-
- Ichikawa N., Ushida S., Kawabata M., Masazumi Y. (1999) Nucleotide sequence of cDNA coding the mitochondrial precursor protein of the ATPase inhibitor from humans. Biosci. Biotechnol. Biochem. 63, 2225–2227 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

