Quantitative analysis of purine nucleotides indicates that purinosomes increase de novo purine biosynthesis
- PMID: 25605736
- PMCID: PMC4358094
- DOI: 10.1074/jbc.M114.628701
Quantitative analysis of purine nucleotides indicates that purinosomes increase de novo purine biosynthesis
Abstract
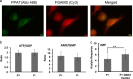
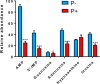
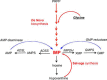
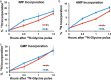
Enzymes in the de novo purine biosynthesis pathway are recruited to form a dynamic metabolic complex referred to as the purinosome. Previous studies have demonstrated that purinosome assembly responds to purine levels in culture medium. Purine-depleted medium or 2-dimethylamino-4,5,6,7-tetrabromo-1H-benzimidazole (DMAT) treatment stimulates the purinosome assembly in HeLa cells. Here, several metabolomic technologies were applied to quantify the static cellular levels of purine nucleotides and measure the de novo biosynthesis rate of IMP, AMP, and GMP. Direct comparison of purinosome-rich cells (cultured in purine-depleted medium) and normal cells showed a 3-fold increase in IMP concentration in purinosome-rich cells and similar levels of AMP, GMP, and ratios of AMP/GMP and ATP/ADP for both. In addition, a higher level of IMP was also observed in HeLa cells treated with DMAT. Furthermore, increases in the de novo IMP/AMP/GMP biosynthetic flux rate under purine-depleted condition were observed. The synthetic enzymes, adenylosuccinate synthase (ADSS) and inosine monophosphate dehydrogenase (IMPDH), downstream of IMP were also shown to be part of the purinosome. Collectively, these results provide further evidence that purinosome assembly is directly related to activated de novo purine biosynthesis, consistent with the functionality of the purinosome.
Keywords: Mass Spectrometry (MS); Metabolic Flux; Metabolomics; Nucleoside/Nucleotide Biosynthesis; Protein Complex; Purine; Purinosome.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures

Similar articles
-
Detecting Purinosome Metabolon Formation with Fluorescence Microscopy.Methods Mol Biol. 2018;1764:279-289. doi: 10.1007/978-1-4939-7759-8_17. Methods Mol Biol. 2018. PMID: 29605921 Free PMC article.
-
Dynamic architecture of the purinosome involved in human de novo purine biosynthesis.Biochemistry. 2015 Jan 27;54(3):870-80. doi: 10.1021/bi501480d. Epub 2015 Jan 15. Biochemistry. 2015. PMID: 25540829 Free PMC article.
-
Hypoxia drives the assembly of the multienzyme purinosome complex.J Biol Chem. 2020 Jul 10;295(28):9551-9566. doi: 10.1074/jbc.RA119.012175. Epub 2020 May 21. J Biol Chem. 2020. PMID: 32439803 Free PMC article.
-
Human de novo purine biosynthesis.Crit Rev Biochem Mol Biol. 2021 Feb;56(1):1-16. doi: 10.1080/10409238.2020.1832438. Epub 2020 Nov 12. Crit Rev Biochem Mol Biol. 2021. PMID: 33179964 Free PMC article. Review.
-
The Purinosome: A Case Study for a Mammalian Metabolon.Annu Rev Biochem. 2022 Jun 21;91:89-106. doi: 10.1146/annurev-biochem-032620-105728. Epub 2022 Mar 23. Annu Rev Biochem. 2022. PMID: 35320684 Free PMC article. Review.
Cited by
-
IMPDH1 retinal variants control filament architecture to tune allosteric regulation.Nat Struct Mol Biol. 2022 Jan;29(1):47-58. doi: 10.1038/s41594-021-00706-2. Epub 2022 Jan 10. Nat Struct Mol Biol. 2022. PMID: 35013599 Free PMC article.
-
Targeting purine metabolism in ovarian cancer.J Ovarian Res. 2022 Aug 13;15(1):93. doi: 10.1186/s13048-022-01022-z. J Ovarian Res. 2022. PMID: 35964092 Free PMC article. Review.
-
Metabolomics and mass spectrometry imaging reveal channeled de novo purine synthesis in cells.Science. 2020 Apr 17;368(6488):283-290. doi: 10.1126/science.aaz6465. Science. 2020. PMID: 32299949 Free PMC article.
-
Biochemical Timekeeping Via Reentrant Phase Transitions.J Mol Biol. 2021 Jun 11;433(12):166794. doi: 10.1016/j.jmb.2020.166794. Epub 2020 Dec 31. J Mol Biol. 2021. PMID: 33387533 Free PMC article. Review.
-
Interplay between mTOR and Purine Metabolism Enzymes and Its Relevant Role in Cancer.Int J Mol Sci. 2024 Jun 19;25(12):6735. doi: 10.3390/ijms25126735. Int J Mol Sci. 2024. PMID: 38928439 Free PMC article. Review.
References
-
- Rowe P. B., McCairns E., Madsen G., Sauer D., Elliott H. (1978) De novo purine synthesis in avian liver: co-purification of the enzymes and properties of the pathway. J. Biol. Chem. 253, 7711–7721 - PubMed
-
- Rowe P. B., Wyngaarden J. B. (1968) Glutamine phosphoribosylpyrophosphate amidotransferase: purification, substructure, amino acid composition, and absorption spectra. J. Biol. Chem. 243, 6373–6383 - PubMed
-
- Smith G. K., Mueller W. T., Wasserman G. F., Taylor W. D., Benkovic S. J. (1980) Characterization of the enzyme complex involving the folate-requiring enzymes of de novo purine biosynthesis. Biochemistry 19, 4313–4321 - PubMed
-
- Wombacher H. (1983) Molecular compartmentation by enzyme cluster formation: a view over current investigations. Mol. Cell. Biochem. 56, 155–164 - PubMed
-
- Srere P. A. (1987) Complexes of sequential metabolic enzymes. Annu. Rev. Biochem. 56, 89–124 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

