Real-time resolution of point mutations that cause phenovariance in mice
- PMID: 25605905
- PMCID: PMC4321302
- DOI: 10.1073/pnas.1423216112
Real-time resolution of point mutations that cause phenovariance in mice
Abstract
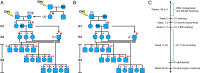
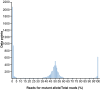
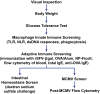
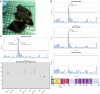
With the wide availability of massively parallel sequencing technologies, genetic mapping has become the rate limiting step in mammalian forward genetics. Here we introduce a method for real-time identification of N-ethyl-N-nitrosourea-induced mutations that cause phenotypes in mice. All mutations are identified by whole exome G1 progenitor sequencing and their zygosity is established in G2/G3 mice before phenotypic assessment. Quantitative and qualitative traits, including lethal effects, in single or multiple combined pedigrees are then analyzed with Linkage Analyzer, a software program that detects significant linkage between individual mutations and aberrant phenotypic scores and presents processed data as Manhattan plots. As multiple alleles of genes are acquired through mutagenesis, pooled "superpedigrees" are created to analyze the effects. Our method is distinguished from conventional forward genetic methods because it permits (1) unbiased declaration of mappable phenotypes, including those that are incompletely penetrant (2), automated identification of causative mutations concurrent with phenotypic screening, without the need to outcross mutant mice to another strain and backcross them, and (3) exclusion of genes not involved in phenotypes of interest. We validated our approach and Linkage Analyzer for the identification of 47 mutations in 45 previously known genes causative for adaptive immune phenotypes; our analysis also implicated 474 genes not previously associated with immune function. The method described here permits forward genetic analysis in mice, limited only by the rates of mutant production and screening.
Keywords: N-ethyl-N-nitrosourea; forward genetics; genetic mapping; massively parallel sequencing; mutagenesis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Schneeberger K. Using next-generation sequencing to isolate mutant genes from forward genetic screens. Nat Rev Genet. 2014;15(10):662–676. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

