Small peptide binding stiffens the ubiquitin-like protein SUMO1
- PMID: 25606684
- PMCID: PMC4302194
- DOI: 10.1016/j.bpj.2014.11.3474
Small peptide binding stiffens the ubiquitin-like protein SUMO1
Abstract
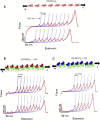
Posttranslational modification by small ubiquitin-like modifiers (SUMOs), known as SUMOylation, is a key regulatory event in many eukaryotic cellular processes in which SUMOs interact with a large number of target proteins. SUMO binding motifs (SBMs) are small peptides derived from these target proteins that interact noncovalently with SUMOs and induce conformational changes. To determine the effect of SBMs on the mechanical properties of SUMO1 (the first member of the human SUMO family), we performed single-molecule force spectroscopy experiments on SUMO1/SBM complexes. The unfolding force of SUMO1 (at a pulling speed of 400 nm/s) increased from ∼ 130 pN to ∼ 170 pN upon binding to SBMs, indicating mechanical stabilization upon complexation. Pulling-speed-dependent experiments and Monte Carlo simulations measured a large decrease in distance to the unfolding transition state for SUMO1 upon SBM binding, which is by far the largest change measured for any ligand binding protein. The stiffness of SUMO1 (measured as a spring constant for the deformation response along the line joining the N- and C-termini) increased upon SBM binding from ∼ 1 N/m to ∼ 3.5 N/m. The relatively higher flexibility of ligand-free SUMO1 might play a role in accessing various conformations before binding to a target.
Copyright © 2015 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures





References
-
- Sanchez-Ruiz J.M. Ligand effects on protein thermodynamic stability. Biophys. Chem. 2007;126:43–49. - PubMed
-
- Hu X., Li H. Force spectroscopy studies on protein-ligand interactions: a single protein mechanics perspective. FEBS Lett. 2014;588:3613–3620. - PubMed
-
- Kotamarthi H.C., Sharma R., Ainavarapu S.R. Multiple unfolding pathways of leucine binding protein (LBP) probed by single-molecule force spectroscopy (SMFS) J. Am. Chem. Soc. 2013;135:14768–14774. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

