Determination of the catalytic mechanism for mitochondrial malate dehydrogenase
- PMID: 25606688
- PMCID: PMC4302198
- DOI: 10.1016/j.bpj.2014.11.3467
Determination of the catalytic mechanism for mitochondrial malate dehydrogenase
Abstract
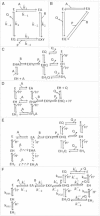
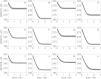
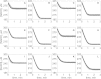
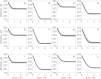
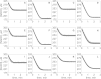
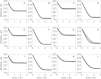
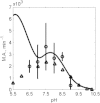
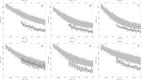
The kinetics of malate dehydrogenase (MDH) catalyzed oxidation/reduction of L-malate/oxaloacetate is pH-dependent due to the proton generated/taken up during the reaction. Previous kinetic studies on the mitochondrial MDH did not yield a consensus kinetic model that explains both substrate and pH dependency of the initial velocity. In this study, we propose, to our knowledge, a new kinetic mechanism to explain kinetic data acquired over a range of pH and substrate concentrations. Progress curves in the forward and reverse reaction directions were obtained under a variety of reactant concentrations to identify associated kinetic parameters. Experiments were conducted at physiologically relevant ionic strength of 0.17 M, pH ranging between 6.5 and 9.0, and at 25 °C. The developed model was built on the prior observation of proton uptake upon binding of NADH to MDH, and that the MDH-catalyzed oxidation of NADH may follow an ordered bi-bi mechanism with NADH/NAD binding to the enzyme first, followed by the binding of oxaloacetate/L-malate. This basic mechanism was expanded to account for additional ionic states to explain the pH dependency of the kinetic behavior, resulting in what we believe to be the first kinetic model explaining both substrate and pH dependency of the reaction velocity.
Copyright © 2015 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures

References
-
- Holbrook J.J., Wolfe R.G. Malate dehydrogenase. X. Fluorescence microtitration studies of D-malate, hydroxymalonate, nicotinamide dinucleotide, and dihydronicotinamide-adenine dinucleotide binding by mitochondrial and supernatant porcine heart enzymes. Biochemistry. 1972;11:2499–2502. - PubMed
-
- Roderick S.L., Banaszak L.J. The conformation of mitochondrial malate dehydrogenase derived from an electron density map at 5.3-Å resolution. J. Biol. Chem. 1983;258:11636–11642. - PubMed
-
- Roderick S.L., Banaszak L.J. The three-dimensional structure of porcine heart mitochondrial malate dehydrogenase at 3.0-Å resolution. J. Biol. Chem. 1986;261:9461–9464. - PubMed
-
- Gleason W.B., Fu Z., Banaszak L. Refined crystal structure of mitochondrial malate dehydrogenase from porcine heart and the consensus structure for dicarboxylic acid oxidoreductases. Biochemistry. 1994;33:2078–2088. - PubMed
-
- Murphey W.H., Kitto G.B., Kaplan N. Malate dehydrogenases. I. A survey of molecular size measured by gel filtration. Biochemistry. 1967;6:603–610. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

