HIV-1 replication and the cellular eukaryotic translation apparatus
- PMID: 25606970
- PMCID: PMC4306834
- DOI: 10.3390/v7010199
HIV-1 replication and the cellular eukaryotic translation apparatus
Abstract
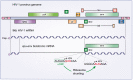
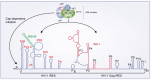
Eukaryotic translation is a complex process composed of three main steps: initiation, elongation, and termination. During infections by RNA- and DNA-viruses, the eukaryotic translation machinery is used to assure optimal viral protein synthesis. Human immunodeficiency virus type I (HIV-1) uses several non-canonical pathways to translate its own proteins, such as leaky scanning, frameshifting, shunt, and cap-independent mechanisms. Moreover, HIV-1 modulates the host translation machinery by targeting key translation factors and overcomes different cellular obstacles that affect protein translation. In this review, we describe how HIV-1 proteins target several components of the eukaryotic translation machinery, which consequently improves viral translation and replication.
Figures



Similar articles
-
HIV-1 translation and its regulation by cellular factors PKR and PACT.Virus Res. 2014 Nov 26;193:65-77. doi: 10.1016/j.virusres.2014.07.014. Epub 2014 Jul 23. Virus Res. 2014. PMID: 25064266 Review.
-
Focus on Translation Initiation of the HIV-1 mRNAs.Int J Mol Sci. 2018 Dec 28;20(1):101. doi: 10.3390/ijms20010101. Int J Mol Sci. 2018. PMID: 30597859 Free PMC article. Review.
-
Dual role of host cell factors in HIV-1 replication: restriction and enhancement of the viral cycle.AIDS Rev. 2010 Apr-Jun;12(2):103-12. AIDS Rev. 2010. PMID: 20571604 Review.
-
Translation initiation is driven by different mechanisms on the HIV-1 and HIV-2 genomic RNAs.Virus Res. 2013 Feb;171(2):366-81. doi: 10.1016/j.virusres.2012.10.006. Epub 2012 Oct 16. Virus Res. 2013. PMID: 23079111 Review.
-
Tinkering with translation: protein synthesis in virus-infected cells.Cold Spring Harb Perspect Biol. 2013 Jan 1;5(1):a012351. doi: 10.1101/cshperspect.a012351. Cold Spring Harb Perspect Biol. 2013. PMID: 23209131 Free PMC article. Review.
Cited by
-
Advances in Continuous Microfluidics-Based Technologies for the Study of HIV Infection.Viruses. 2020 Sep 4;12(9):982. doi: 10.3390/v12090982. Viruses. 2020. PMID: 32899657 Free PMC article. Review.
-
Viral and Host Factors Regulating HIV-1 Envelope Protein Trafficking and Particle Incorporation.Viruses. 2022 Aug 5;14(8):1729. doi: 10.3390/v14081729. Viruses. 2022. PMID: 36016351 Free PMC article. Review.
-
Death of a dogma: eukaryotic mRNAs can code for more than one protein.Nucleic Acids Res. 2016 Jan 8;44(1):14-23. doi: 10.1093/nar/gkv1218. Epub 2015 Nov 17. Nucleic Acids Res. 2016. PMID: 26578573 Free PMC article. Review.
-
Structural domains within the HIV-1 mRNA and the ribosomal protein S25 influence cap-independent translation initiation.FEBS J. 2016 Jul;283(13):2508-27. doi: 10.1111/febs.13756. Epub 2016 Jun 10. FEBS J. 2016. PMID: 27191820 Free PMC article.
-
Interactions between HIV proteins and host restriction factors: implications for potential therapeutic intervention in HIV infection.Front Immunol. 2024 Aug 16;15:1390650. doi: 10.3389/fimmu.2024.1390650. eCollection 2024. Front Immunol. 2024. PMID: 39221250 Free PMC article. Review.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

