Usutu virus: an emerging flavivirus in Europe
- PMID: 25606971
- PMCID: PMC4306835
- DOI: 10.3390/v7010219
Usutu virus: an emerging flavivirus in Europe
Abstract
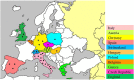
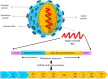
Usutu virus (USUV) is an African mosquito-borne flavivirus belonging to the Japanese encephalitis virus serocomplex. USUV is closely related to Murray Valley encephalitis virus, Japanese encephalitis virus, and West Nile virus. USUV was discovered in South Africa in 1959. In Europe, the first true demonstration of circulation of USUV was reported in Austria in 2001 with a significant die-off of Eurasian blackbirds. In the subsequent years, USUV expanded to neighboring countries, including Italy, Germany, Spain, Hungary, Switzerland, Poland, England, Czech Republic, Greece, and Belgium, where it caused unusual mortality in birds. In 2009, the first two human cases of USUV infection in Europe have been reported in Italy, causing meningoencephalitis in immunocompromised patients. This review describes USUV in terms of its life cycle, USUV surveillance from Africa to Europe, human cases, its cellular tropism and pathogenesis, its genetic relationship with other flaviviruses, genetic diversity among USUV strains, its diagnosis, and a discussion of the potential future threat to Asian countries.
Figures


References
-
- Karabatsos N. International Catalogue of Arboviruses Including Certain other Viruses of Vertebrates. 3rd ed. The American Society of Tropical Medicine and Hygiene; San Antonio, TX, USA: 1985. Usutu virus; pp. 1059–1060. - PubMed
-
- Calisher C.H., Gould E.A. Taxonomy of the virus family Flaviviridae. Adv. Virus Res. 2003;59:1–19. - PubMed
-
- Woodall J.P. The viruses isolated from arthropods at the East African virus research institute in the 26 years ending December 1963. Proc. E. Afr. Acad. 1964;2:141–146.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

