Common genetic variants influence human subcortical brain structures
- PMID: 25607358
- PMCID: PMC4393366
- DOI: 10.1038/nature14101
Common genetic variants influence human subcortical brain structures
Abstract
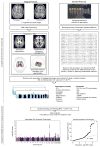
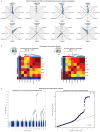
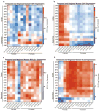
The highly complex structure of the human brain is strongly shaped by genetic influences. Subcortical brain regions form circuits with cortical areas to coordinate movement, learning, memory and motivation, and altered circuits can lead to abnormal behaviour and disease. To investigate how common genetic variants affect the structure of these brain regions, here we conduct genome-wide association studies of the volumes of seven subcortical regions and the intracranial volume derived from magnetic resonance images of 30,717 individuals from 50 cohorts. We identify five novel genetic variants influencing the volumes of the putamen and caudate nucleus. We also find stronger evidence for three loci with previously established influences on hippocampal volume and intracranial volume. These variants show specific volumetric effects on brain structures rather than global effects across structures. The strongest effects were found for the putamen, where a novel intergenic locus with replicable influence on volume (rs945270; P = 1.08 × 10(-33); 0.52% variance explained) showed evidence of altering the expression of the KTN1 gene in both brain and blood tissue. Variants influencing putamen volume clustered near developmental genes that regulate apoptosis, axon guidance and vesicle transport. Identification of these genetic variants provides insight into the causes of variability in human brain development, and may help to determine mechanisms of neuropsychiatric dysfunction.
Conflict of interest statement
The authors declare no competing financial interests.
Figures






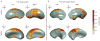

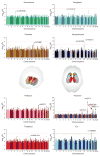
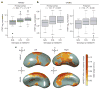
References
-
- Poldrack RA, et al. Interactive memory systems in the human brain. Nature. 2001;414:546–550. - PubMed
MeSH terms
Substances
Grants and funding
- P41 EB015922/EB/NIBIB NIH HHS/United States
- R00 LM011384/LM/NLM NIH HHS/United States
- U24 AG021886/AG/NIA NIH HHS/United States
- G0701120/MRC_/Medical Research Council/United Kingdom
- G0900908/MRC_/Medical Research Council/United Kingdom
- BB/F019394/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- R01 NS017950/NS/NINDS NIH HHS/United States
- R01 AG019771/AG/NIA NIH HHS/United States
- RF1 AG041915/AG/NIA NIH HHS/United States
- G0802462/MRC_/Medical Research Council/United Kingdom
- K99 MH101367/MH/NIMH NIH HHS/United States
- K99 LM011384/LM/NLM NIH HHS/United States
- ETM/84/CSO_/Chief Scientist Office/United Kingdom
- R01 AG040060/AG/NIA NIH HHS/United States
- 104036/WT_/Wellcome Trust/United Kingdom
- P50 AG005146/AG/NIA NIH HHS/United States
- R01 EB015611/EB/NIBIB NIH HHS/United States
- P30 AG062421/AG/NIA NIH HHS/United States
- R01 EB005846/EB/NIBIB NIH HHS/United States
- T32 NS048004/NS/NINDS NIH HHS/United States
- MR/L016400/1/MRC_/Medical Research Council/United Kingdom
- R01 EB006841/EB/NIBIB NIH HHS/United States
- P30 AG013846/AG/NIA NIH HHS/United States
- G1001245/MRC_/Medical Research Council/United Kingdom
- MR/K026992/1/MRC_/Medical Research Council/United Kingdom
- MR/N000390/1/MRC_/Medical Research Council/United Kingdom
- UL1 TR001108/TR/NCATS NIH HHS/United States
- U54 EB020403/EB/NIBIB NIH HHS/United States
- R01 AG008122/AG/NIA NIH HHS/United States
- P50 AG005134/AG/NIA NIH HHS/United States
- 100309/WT_/Wellcome Trust/United Kingdom
- R01 AG033193/AG/NIA NIH HHS/United States
- K01 MH099232/MH/NIMH NIH HHS/United States
- P50 AG005133/AG/NIA NIH HHS/United States
- G0901858/MRC_/Medical Research Council/United Kingdom
- P30 AG010129/AG/NIA NIH HHS/United States
- UL1 TR001120/TR/NCATS NIH HHS/United States
- G0700704/MRC_/Medical Research Council/United Kingdom
- R01 EB020407/EB/NIBIB NIH HHS/United States
- R01 MH090553/MH/NIMH NIH HHS/United States
- U01 AG049505/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

