Long ncRNA expression associates with tissue-specific enhancers
- PMID: 25607649
- PMCID: PMC4614992
- DOI: 10.4161/15384101.2014.977641
Long ncRNA expression associates with tissue-specific enhancers
Abstract
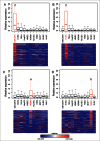
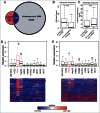
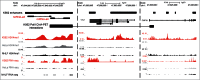
Long non-coding RNAs (ncRNA) have recently been demonstrated to be expressed from a subset of enhancers and to be required for the distant regulation of gene expression. Several approaches to predict enhancers have been developed based on various chromatin marks and occupancy of enhancer-binding proteins. Despite the rapid advances in the field, no consensus how to define tissue specific enhancers yet exists. Here, we identify 2,695 long ncRNAs annotated by ENCODE (corresponding to 28% of all ENCODE annotated long ncRNAs) that overlap tissue-specific enhancers. We use a recently developed algorithm to predict tissue-specific enhancers, PreSTIGE, that is based on the H3K4me1 mark and tissue specific expression of mRNAs. The expression of the long ncRNAs overlapping enhancers is significantly higher when the enhancer is predicted as active in a specific cell line, suggesting a general interdependency of active enhancers and expression of long ncRNAs. This dependency is not identified using previous enhancer prediction algorithms that do not account for expression of their downstream targets. The predicted enhancers that overlap annotated long ncRNAs generally have a lower ratio of H3K4me1 to H3K4me3, suggesting that enhancers expressing long ncRNAs might be associated with specific epigenetic marks. In conclusion, we demonstrate the tissue-specific predictive power of PreSTIGE and provide evidence for thousands of long ncRNAs that are expressed from active tissue-specific enhancers, suggesting a particularly important functional relationship between long ncRNAs and enhancer activity in determining tissue-specific gene expression.
Keywords: Enhancer; activating long non-coding RNA; enhancer prediction; long non-coding RNA; tissue-specific enhancer.
Figures

Comment in
-
A PreSTIGEous use of LncRNAs to predict enhancers.Cell Cycle. 2015;14(11):1619-20. doi: 10.1080/15384101.2015.1032650. Cell Cycle. 2015. PMID: 25928187 Free PMC article. No abstract available.
References
-
- Calo E, Wysocka J. Modification of enhancer chromatin: what, how, and why? Mol Cell 2013; 49:825-37; PMID:23473601; http://dx.doi.org/10.1016/j.molcel.2013.01.038 - DOI - PMC - PubMed
-
- De Santa F, Barozzi I, Mietton F, Ghisletti S, Polletti S, Tusi BK, Muller H, Ragoussis J, Wei CL, Natoli G. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol 2010; 8:e1000384; PMID:20485488; http://dx.doi.org/10.1371/journal.pbio.1000384 - DOI - PMC - PubMed
-
- Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, et al. . Long noncoding RNAs with enhancer-like function in human cells. Cell 2010; 143:46-58; PMID:20887892; http://dx.doi.org/10.1016/j.cell.2010.09.001 - DOI - PMC - PubMed
-
- Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, et al. . Widespread transcription at neuronal activity-regulated enhancers. Nature 2010; 465:182-7; PMID:20393465; http://dx.doi.org/10.1038/nature09033 - DOI - PMC - PubMed
-
- Orom UA, Shiekhattar R. Long noncoding RNAs usher in a new era in the biology of enhancers. Cell 2013; 154:1190-3; PMID:24034243; http://dx.doi.org/10.1016/j.cell.2013.08.028 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
