The adhesion G protein-coupled receptor GPR56 is a cell-autonomous regulator of oligodendrocyte development
- PMID: 25607655
- PMCID: PMC4302951
- DOI: 10.1038/ncomms7121
The adhesion G protein-coupled receptor GPR56 is a cell-autonomous regulator of oligodendrocyte development
Abstract
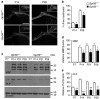
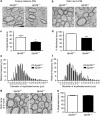
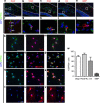
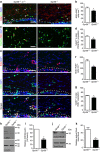
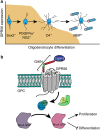
Mutations in GPR56, a member of the adhesion G protein-coupled receptor family, cause a human brain malformation called bilateral frontoparietal polymicrogyria (BFPP). Magnetic resonance imaging (MRI) of BFPP brains reveals myelination defects in addition to brain malformation. However, the cellular role of GPR56 in oligodendrocyte development remains unknown. Here, we demonstrate that loss of Gpr56 leads to hypomyelination of the central nervous system in mice. GPR56 levels are abundant throughout early stages of oligodendrocyte development, but are downregulated in myelinating oligodendrocytes. Gpr56-knockout mice manifest with decreased oligodendrocyte precursor cell (OPC) proliferation and diminished levels of active RhoA, leading to fewer mature oligodendrocytes and a reduced number of myelinated axons in the corpus callosum and optic nerves. Conditional ablation of Gpr56 in OPCs leads to a reduced number of mature oligodendrocytes as seen in constitutive knockout of Gpr56. Together, our data define GPR56 as a cell-autonomous regulator of oligodendrocyte development.
Figures



References
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

