Experimentally-derived fibroblast gene signatures identify molecular pathways associated with distinct subsets of systemic sclerosis patients in three independent cohorts
- PMID: 25607805
- PMCID: PMC4301872
- DOI: 10.1371/journal.pone.0114017
Experimentally-derived fibroblast gene signatures identify molecular pathways associated with distinct subsets of systemic sclerosis patients in three independent cohorts
Abstract
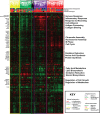
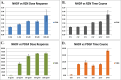
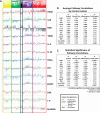
Genome-wide expression profiling in systemic sclerosis (SSc) has identified four 'intrinsic' subsets of disease (fibroproliferative, inflammatory, limited, and normal-like), each of which shows deregulation of distinct signaling pathways; however, the full set of pathways contributing to this differential gene expression has not been fully elucidated. Here we examine experimentally derived gene expression signatures in dermal fibroblasts for thirteen different signaling pathways implicated in SSc pathogenesis. These data show distinct and overlapping sets of genes induced by each pathway, allowing for a better understanding of the molecular relationship between profibrotic and immune signaling networks. Pathway-specific gene signatures were analyzed across a compendium of microarray datasets consisting of skin biopsies from three independent cohorts representing 80 SSc patients, 4 morphea, and 26 controls. IFNα signaling showed a strong association with early disease, while TGFβ signaling spanned the fibroproliferative and inflammatory subsets, was associated with worse MRSS, and was higher in lesional than non-lesional skin. The fibroproliferative subset was most strongly associated with PDGF signaling, while the inflammatory subset demonstrated strong activation of innate immune pathways including TLR signaling upstream of NF-κB. The limited and normal-like subsets did not show associations with fibrotic and inflammatory mediators such as TGFβ and TNFα. The normal-like subset showed high expression of genes associated with lipid signaling, which was absent in the inflammatory and limited subsets. Together, these data suggest a model by which IFNα is involved in early disease pathology, and disease severity is associated with active TGFβ signaling.
Conflict of interest statement
Figures

References
-
- LeRoy EC, Black C, Fleischmajer R, Jablonska S, Krieg T, et al. (1988) Scleroderma (systemic sclerosis): classification, subsets and pathogenesis. J Rheumatol 15: 202 - PubMed
Publication types
MeSH terms
Grants and funding
- U01 AR055063/AR/NIAMS NIH HHS/United States
- P30 CA023108/CA/NCI NIH HHS/United States
- T32 GM008704/GM/NIGMS NIH HHS/United States
- T32GM008704/GM/NIGMS NIH HHS/United States
- P30 AR061271/AR/NIAMS NIH HHS/United States
- R25 CA134286-01/CA/NCI NIH HHS/United States
- R25 CA134286/CA/NCI NIH HHS/United States
- P30AR061271/AR/NIAMS NIH HHS/United States
- U01AR055063/AR/NIAMS NIH HHS/United States
- P50AR060780/AR/NIAMS NIH HHS/United States
- P50 AR060780/AR/NIAMS NIH HHS/United States
- K23 AR059763/AR/NIAMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

