Evolutionary relationships among barley and Arabidopsis core circadian clock and clock-associated genes
- PMID: 25608480
- PMCID: PMC4320304
- DOI: 10.1007/s00239-015-9665-0
Evolutionary relationships among barley and Arabidopsis core circadian clock and clock-associated genes
Abstract
The circadian clock regulates a multitude of plant developmental and metabolic processes. In crop species, it contributes significantly to plant performance and productivity and to the adaptation and geographical range over which crops can be grown. To understand the clock in barley and how it relates to the components in the Arabidopsis thaliana clock, we have performed a systematic analysis of core circadian clock and clock-associated genes in barley, Arabidopsis and another eight species including tomato, potato, a range of monocotyledonous species and the moss, Physcomitrella patens. We have identified orthologues and paralogues of Arabidopsis genes which are conserved in all species, monocot/dicot differences, species-specific differences and variation in gene copy number (e.g. gene duplications among the various species). We propose that the common ancestor of barley and Arabidopsis had two-thirds of the key clock components identified in Arabidopsis prior to the separation of the monocot/dicot groups. After this separation, multiple independent gene duplication events took place in both monocot and dicot ancestors.
Figures



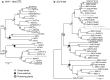

References
-
- Boxall SF, Foster JM, Bohnert HJ, Cushman JC, Nimmo HG, Hartwell J. Conservation and divergence of circadian clock operation in a stress-inducible Crassulacean acid metabolism species reveals clock compensation against stress. Plant Physiol. 2005;137:969–982. doi: 10.1104/pp.104.054577. - DOI - PMC - PubMed
-
- Campoli C, Shtaya M, Davis SJ, von Korff M. Expression conservation within the circadian clock of a monocot: natural variation at barley Ppd-H1 affects circadian expression of flowering time genes, but not clock orthologs. BMC Plant Biol. 2012;12:97. doi: 10.1186/1471-2229-12-97. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

