Experimental confirmation of a whole set of tRNA molecules in two archaeal species
- PMID: 25608653
- PMCID: PMC4307357
- DOI: 10.3390/ijms16012187
Experimental confirmation of a whole set of tRNA molecules in two archaeal species
Abstract
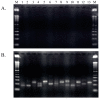
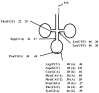
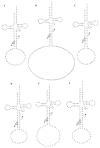
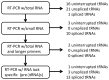
Based on the genomic sequences for most archaeal species, only one tRNA gene (isodecoder) is predicted for each triplet codon. This observation promotes analysis of a whole set of tRNA molecules and actual splicing patterns of interrupted tRNA in one organism. The entire genomic sequences of two Creanarchaeota, Aeropyrum pernix and Sulfolobus tokodaii, were determined approximately 15 years ago. In these genome datasets, 47 and 46 tRNA genes were detected, respectively. Among them, 14 and 24 genes, respectively, were predicted to be interrupted tRNA genes. To confirm the actual transcription of these predicted tRNA genes and identify the actual splicing patterns of the predicted interrupted tRNA molecules, RNA samples were prepared from each archaeal species and used to synthesize cDNA molecules with tRNA sequence-specific primers. Comparison of the nucleotide sequences of cDNA clones representing unspliced and spliced forms of interrupted tRNA molecules indicated that some introns were located at positions other than one base 3' from anticodon region and that bulge-helix-bulge structures were detected around the actual splicing sites in each interrupted tRNA molecule. Whole-set analyses of tRNA molecules revealed that the archaeal tRNA splicing mechanism may be essential for efficient splicing of all tRNAs produced from interrupted tRNA genes in these archaea.
Figures





References
-
- Szekely E., Belford H.G., Greer C.L. Intron sequence and structure requirements for tRNA splicing in Saccharomyces cerevisiae. J. Biol. Chem. 1988;263:13839–13847. - PubMed
-
- Thompson L.D., Daniels C.J. A tRNA(Trp) intron endonuclease from Halobacterium volcanii. Unique substrate recognition properties. J. Biol. Chem. 1988;263:17951–17959. - PubMed
-
- Thompson L.D., Daniels C.J. Recognition of exon-intron boundaries by the Halobacterium volcanii tRNA intron endonuclease. J. Biol. Chem. 1990;265:18104–18111. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

