Repeat instability during DNA repair: Insights from model systems
- PMID: 25608779
- PMCID: PMC4454471
- DOI: 10.3109/10409238.2014.999192
Repeat instability during DNA repair: Insights from model systems
Abstract
The expansion of repeated sequences is the cause of over 30 inherited genetic diseases, including Huntington disease, myotonic dystrophy (types 1 and 2), fragile X syndrome, many spinocerebellar ataxias, and some cases of amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD). Repeat expansions are dynamic, and disease inheritance and progression are influenced by the size and the rate of expansion. Thus, an understanding of the various cellular mechanisms that cooperate to control or promote repeat expansions is of interest to human health. In addition, the study of repeat expansion and contraction mechanisms has provided insight into how repair pathways operate in the context of structure-forming DNA, as well as insights into non-canonical roles for repair proteins. Here we review the mechanisms of repeat instability, with a special emphasis on the knowledge gained from the various model systems that have been developed to study this topic. We cover the repair pathways and proteins that operate to maintain genome stability, or in some cases cause instability, and the cross-talk and interactions between them.
Keywords: Chromosome fragility; DNA damage checkpoint; DNA structure; recombination; repair; replication; structure-specific helicases; trinucleotide repeat expansion.
Figures

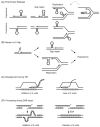
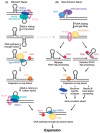
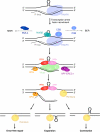
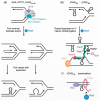

References
-
- Balakumaran BS, Freudenreich CH, Zakian VA. CGG/CCG repeats exhibit orientation-dependent instability and orientation-independent fragility in Saccharomyces cerevisiae. Hum Mol Genet. 2000;9:93–100. - PubMed
-
- Belotserkovskii BP, Mirkin SM, Hanawalt PC. DNA sequences that interfere with transcription: implications for genome function and stability. Chem Rev. 2013;113:8620–37. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
