Quantification of cell identity from single-cell gene expression profiles
- PMID: 25608970
- PMCID: PMC4354993
- DOI: 10.1186/s13059-015-0580-x
Quantification of cell identity from single-cell gene expression profiles
Abstract
The definition of cell identity is a central problem in biology. While single-cell RNA-seq provides a wealth of information regarding cell states, better methods are needed to map their identity, especially during developmental transitions. Here, we use repositories of cell type-specific transcriptomes to quantify identities from single-cell RNA-seq profiles, accurately classifying cells from Arabidopsis root tips and human glioblastoma tumors. We apply our approach to single cells captured from regenerating roots following tip excision. Our technique exposes a previously uncharacterized transient collapse of identity distant from the injury site, demonstrating the biological relevance of a quantitative cell identity index.
Figures
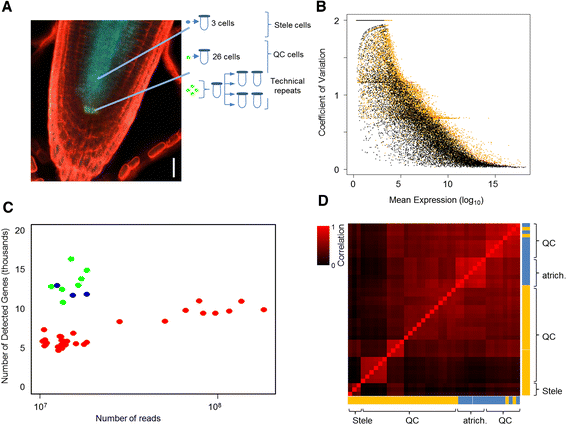
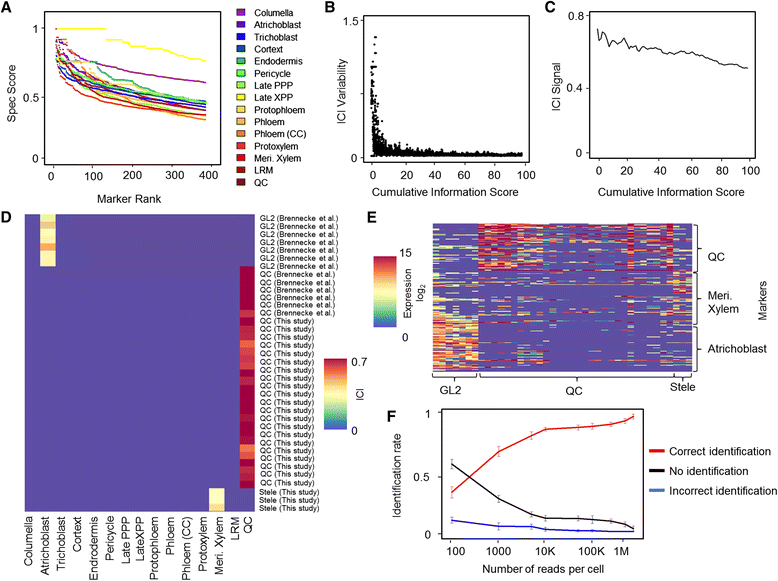
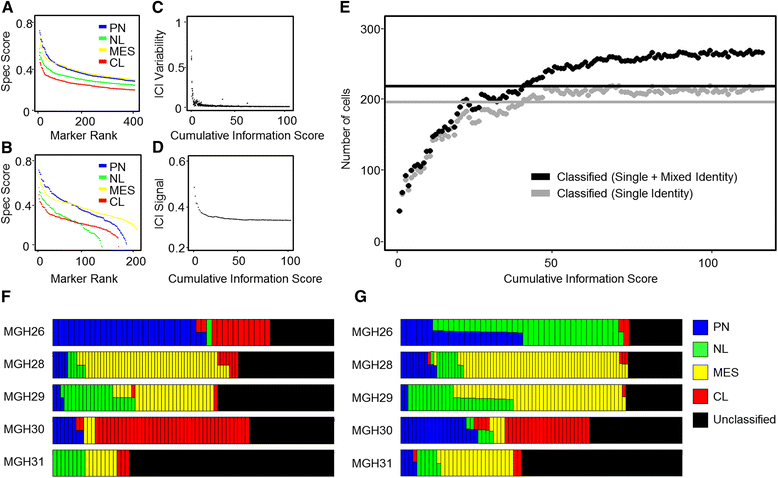
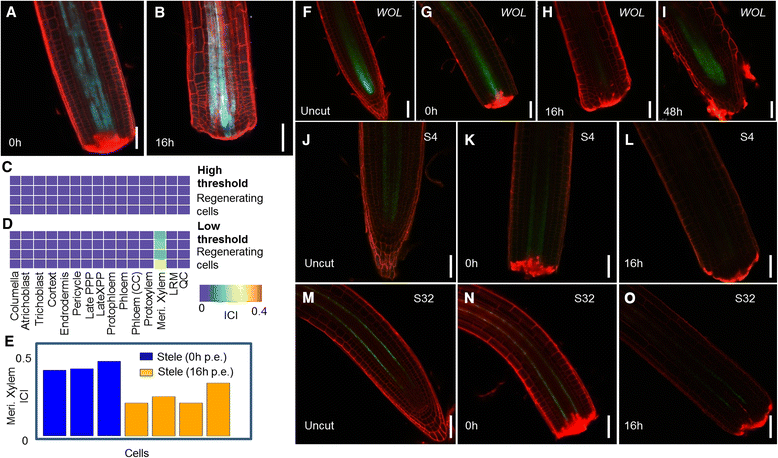
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

