Early B-cell factor 3 (EBF3) is a novel tumor suppressor gene with promoter hypermethylation in pediatric acute myeloid leukemia
- PMID: 25609158
- PMCID: PMC4311429
- DOI: 10.1186/s13046-014-0118-1
Early B-cell factor 3 (EBF3) is a novel tumor suppressor gene with promoter hypermethylation in pediatric acute myeloid leukemia
Abstract
Background: Pediatric acute myeloid leukemia (AML) comprises up to 20% of all childhood leukemia. Recent research shows that aberrant DNA methylation patterning may play a role in leukemogenesis. The epigenetic silencing of the EBF3 locus is very frequent in glioblastoma. However, the expression profiles and molecular function of EBF3 in pediatric AML is still unclear.
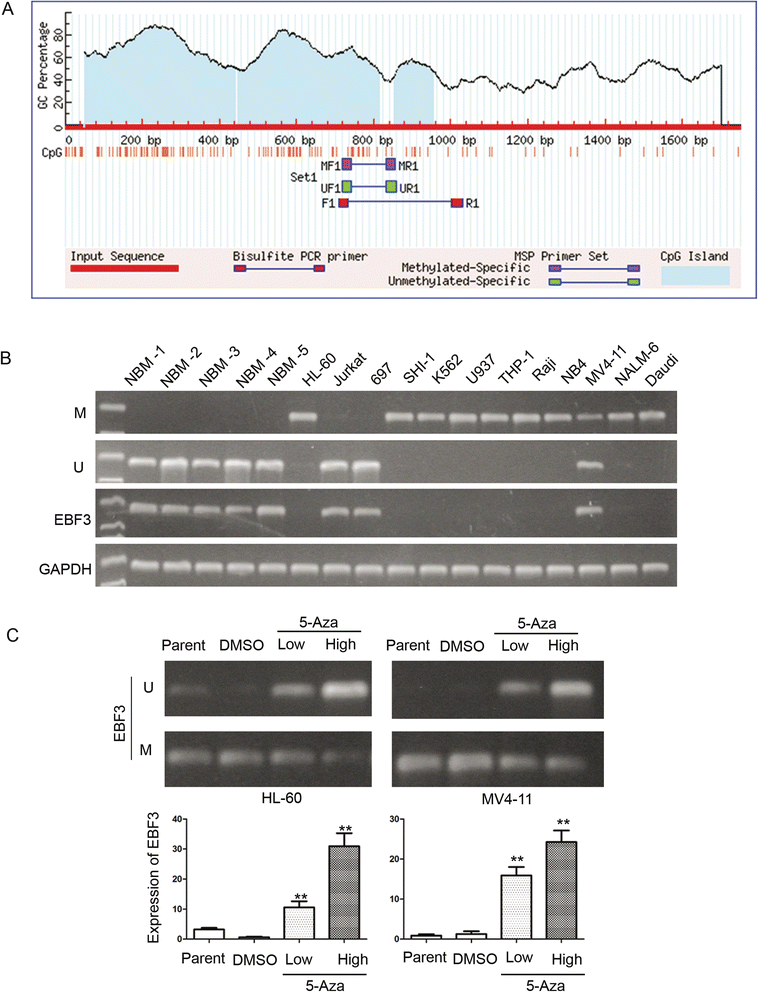
Methods: Twelve human acute leukemia cell lines, 105 pediatric AML samples and 30 normal bone marrow/idiopathic thrombocytopenic purpura (NBM/ITP) control samples were analyzed. Transcriptional level of EBF3 was evaluated by semi-quantitative and real-time PCR. EBF3 methylation status was determined by methylation specific PCR (MSP) and bisulfite genomic sequencing (BGS). The molecular mechanism of EBF3 was investigated by apoptosis assays and PCR array analysis.
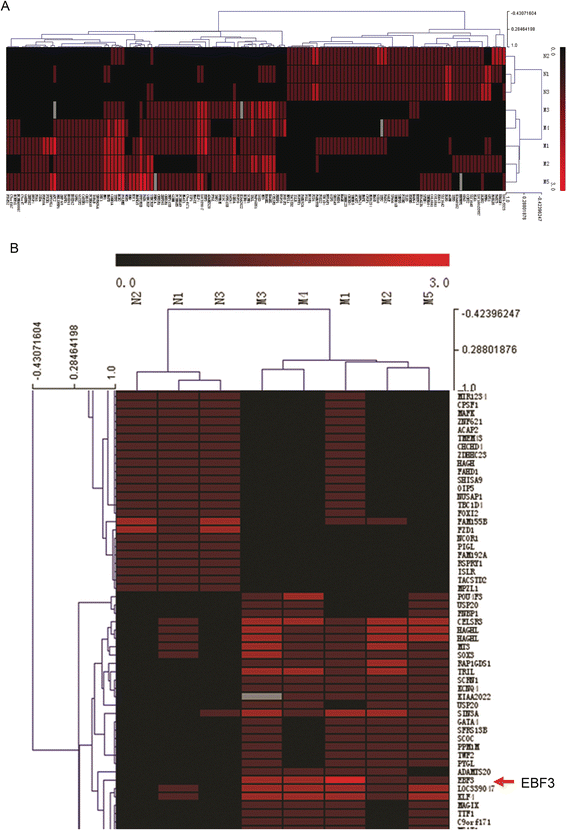
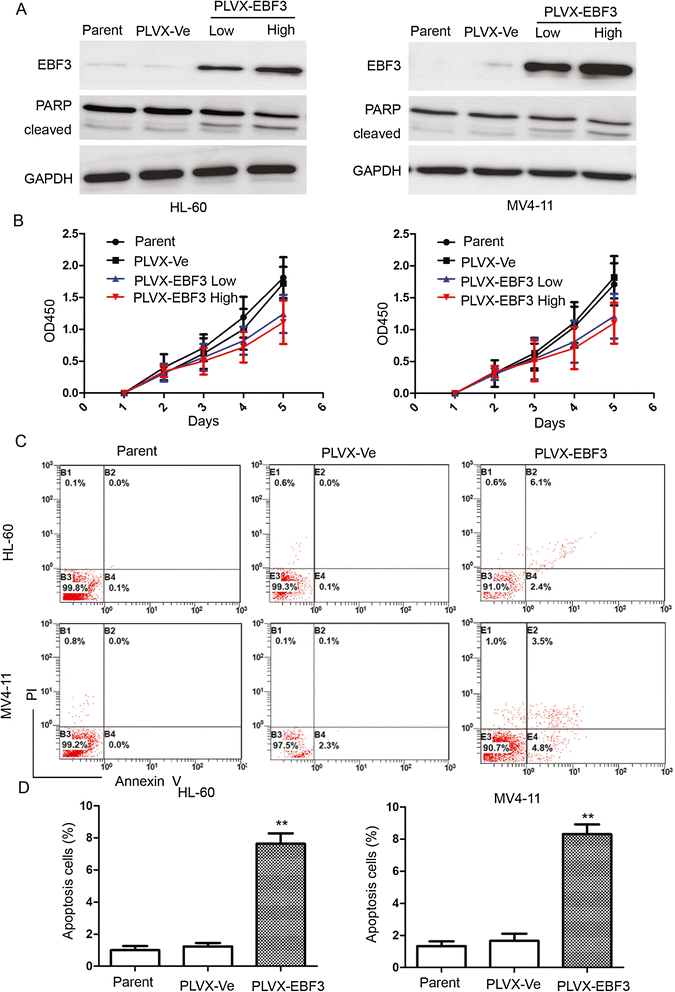
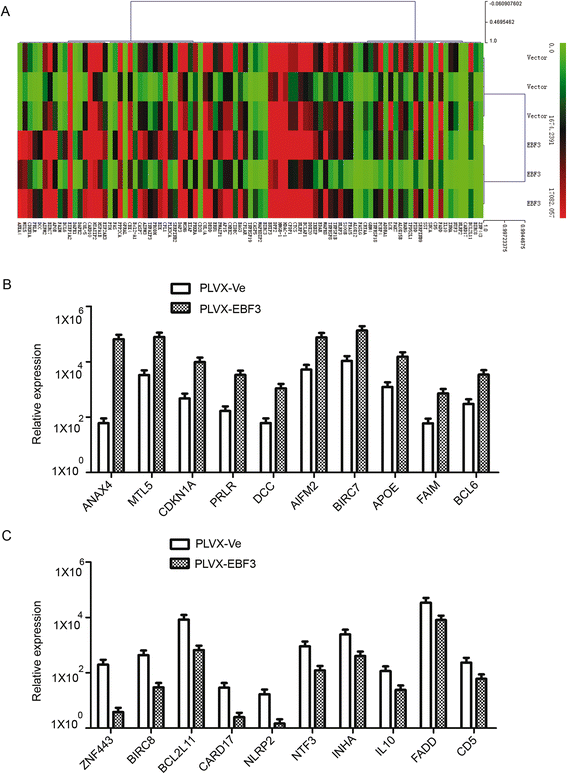
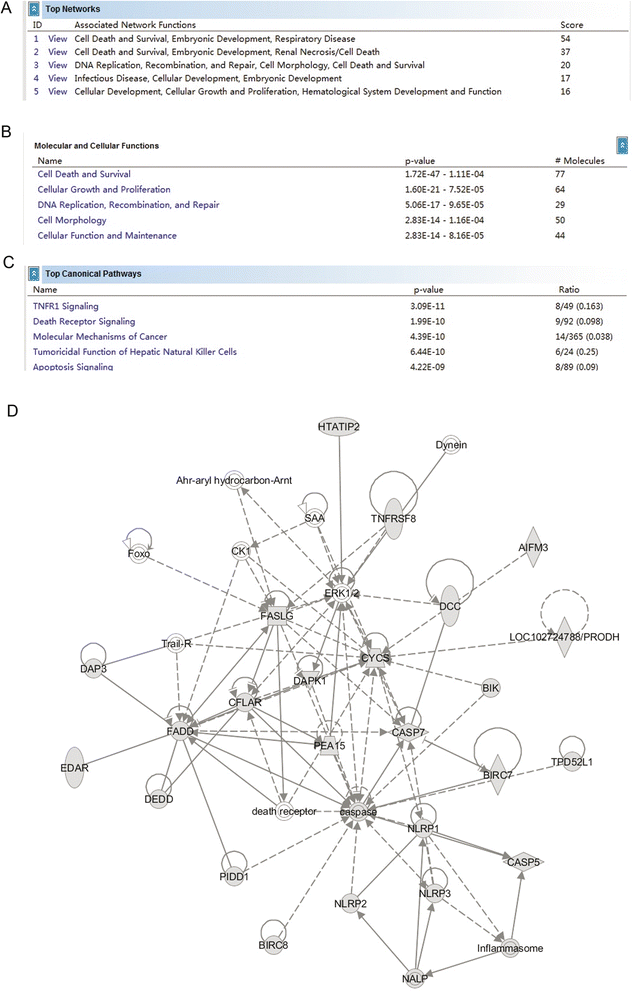
Results: EBF3 promoter was hypermethylated in 10/12 leukemia cell lines. Aberrant EBF3 methylation was observed in 42.9% (45/105) of the pediatric AML samples using MSP analysis, and the BGS results confirmed promoter methylation. EBF3 expression was decreased in the AML samples compared with control. Methylated samples revealed similar survival outcomes by Kaplan-Meier survival analysis. EBF3 overexpression significantly inhibited cell proliferation and increased apoptosis. Real-time PCR array analysis revealed 93 dysregulated genes possibly implicated in the apoptosis of EBF3-induced AML cells.
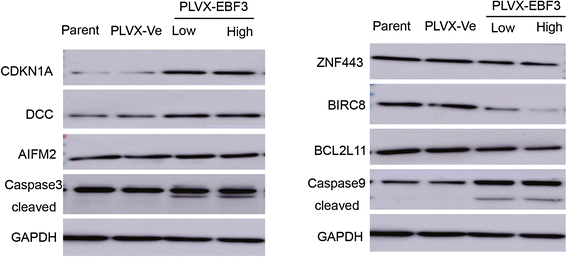
Conclusion: In this study, we firstly identified epigenetic inactivation of EBF3 in both AML cell lines and pediatric AML samples for the first time. Our findings also showed for the first time that transcriptional overexpression of EBF3 could inhibit proliferation and induce apoptosis in AML cells. We identified 93 dysregulated apoptosis-related genes in EBF3-overexpressing, including DCC, AIFM2 and DAPK1. Most of these genes have never been related with EBF3 over expression. These results may provide new insights into the molecular mechanism of EBF3-induced apoptosis; however, further research will be required to determine the underlying details. Our findings suggest that EBF3 may act as a putative tumor suppressor gene in pediatric AML.
Figures



References
-
- Appelbaum FR, Baer MR, Carabasi MH, Coutre SE, Erba HP, Estey E, et al. NCCN practice guidelines for acute myelogenous leukemia. Oncology (Williston Park) 2000;14:53–61. - PubMed
-
- Sanz GF, Sanz MA, Vallespi T, Canizo MC, Torrabadella M, Garcia S, et al. Two regression models and a scoring system for predicting survival and planning treatment in myelodysplastic syndromes: a multivariate analysis of prognostic factors in 370 patients. Blood. 1989;74:395–408. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

