An integrated phylogenomic approach toward pinpointing the origin of mitochondria
- PMID: 25609566
- PMCID: PMC4302308
- DOI: 10.1038/srep07949
An integrated phylogenomic approach toward pinpointing the origin of mitochondria
Abstract
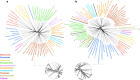
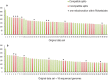
Overwhelming evidence supports the endosymbiosis theory that mitochondria originated once from the Alphaproteobacteria. However, its exact position in the tree of life remains highly debated. This is because systematic errors, including biased taxonomic sampling, high evolutionary rates and sequence composition bias have long plagued the mitochondrial phylogenetics. In this study, we address this issue by 1) increasing the taxonomic representation of alphaproteobacterial genomes by sequencing 18 phylogenetically novel species. They include 5 Rickettsiales and 4 Rhodospirillales, two orders that have shown close affiliations with mitochondria previously, 2) using a set of 29 slowly evolving mitochondria-derived nuclear genes that are less biased than mitochondria-encoded genes as the alternative "well behaved" markers for phylogenetic analysis, 3) applying site heterogeneous mixture models that account for the sequence composition bias. With the integrated phylogenomic approach, we are able to for the first time place mitochondria unequivocally within the Rickettsiales order, as a sister clade to the Rickettsiaceae and Anaplasmataceae families, all subtended by the Holosporaceae family. Our results suggest that mitochondria most likely originated from a Rickettsiales endosymbiont already residing in the host, but not from the distantly related free-living Pelagibacter and Rhodospirillales.
Figures




References
-
- Lang B. F., Seif E., Gray M. W., O'Kelly C. J. & Burger G. A comparative genomics approach to the evolution of eukaryotes and their mitochondria. J. Eukaryot. Microbiol. 46, 320–326 (1999). - PubMed
-
- Gabaldon T. & Huynen M. A. Reconstruction of the proto-mitochondrial metabolism. Science 301, 609 (2003). - PubMed
-
- Gray M. W., Burger G. & Lang B. F. Mitochondrial evolution. Science 283, 1476–1481 (1999). - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

