Large-scale extraction of brain connectivity from the neuroscientific literature
- PMID: 25609795
- PMCID: PMC4426844
- DOI: 10.1093/bioinformatics/btv025
Large-scale extraction of brain connectivity from the neuroscientific literature
Abstract
Motivation: In neuroscience, as in many other scientific domains, the primary form of knowledge dissemination is through published articles. One challenge for modern neuroinformatics is finding methods to make the knowledge from the tremendous backlog of publications accessible for search, analysis and the integration of such data into computational models. A key example of this is metascale brain connectivity, where results are not reported in a normalized repository. Instead, these experimental results are published in natural language, scattered among individual scientific publications. This lack of normalization and centralization hinders the large-scale integration of brain connectivity results. In this article, we present text-mining models to extract and aggregate brain connectivity results from 13.2 million PubMed abstracts and 630 216 full-text publications related to neuroscience. The brain regions are identified with three different named entity recognizers (NERs) and then normalized against two atlases: the Allen Brain Atlas (ABA) and the atlas from the Brain Architecture Management System (BAMS). We then use three different extractors to assess inter-region connectivity.
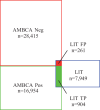
Results: NERs and connectivity extractors are evaluated against a manually annotated corpus. The complete in litero extraction models are also evaluated against in vivo connectivity data from ABA with an estimated precision of 78%. The resulting database contains over 4 million brain region mentions and over 100 000 (ABA) and 122 000 (BAMS) potential brain region connections. This database drastically accelerates connectivity literature review, by providing a centralized repository of connectivity data to neuroscientists.
© The Author 2015. Published by Oxford University Press.
Figures


References
-
- Bowden D., Dubach M. (2003) NeuroNames 2002. Neuroinformatics , 1, 43–59. - PubMed
-
- Burns G., et al. (2008) Intelligent approaches to mining the primary research literature: Techniques, systems, and examples. In: Computational Intelligence in Medical Informatics . Vol. 85 Springer, Berlin, pp. 17–50.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

