Structure of the extracellular domain of matrix protein 2 of influenza A virus in complex with a protective monoclonal antibody
- PMID: 25609808
- PMCID: PMC4403401
- DOI: 10.1128/JVI.02576-14
Structure of the extracellular domain of matrix protein 2 of influenza A virus in complex with a protective monoclonal antibody
Abstract
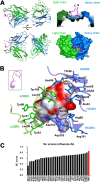
The extracellular domain of influenza A virus matrix protein 2 (M2e) is conserved and is being evaluated as a quasiuniversal influenza A vaccine candidate. We describe the crystal structure at 1.6 Å resolution of M2e in complex with the Fab fragment of an M2e-specific monoclonal antibody that protects against influenza A virus challenge. This antibody binds M2 expressed on the surfaces of cells infected with influenza A virus. Five out of six complementary determining regions interact with M2e, and three highly conserved M2e residues are critical for this interaction. In this complex, M2e adopts a compact U-shaped conformation stabilized in the center by the highly conserved tryptophan residue in M2e. This is the first description of the three-dimensional structure of M2e.
Importance: M2e of influenza A is under investigation as a universal influenza A vaccine, but its three-dimensional structure is unknown. We describe the structure of M2e stabilized with an M2e-specific monoclonal antibody that recognizes natural M2. We found that the conserved tryptophan is positioned in the center of the U-shaped structure of M2e and stabilizes its conformation. The structure also explains why previously reported in vivo escape viruses, selected with a similar monoclonal antibody, carried proline residue substitutions at position 10 in M2.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures


References
-
- Pinto LH, Dieckmann GR, Gandhi CS, Papworth CG, Braman J, Shaughnessy MA, Lear JD, Lamb RA, DeGrado WF. 1997. A functionally defined model for the M2 proton channel of influenza A virus suggests a mechanism for its ion selectivity. Proc Natl Acad Sci U S A 94:11301–11306. doi:10.1073/pnas.94.21.11301. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources

