Plant membrane assays with cytokinin receptors underpin the unique role of free cytokinin bases as biologically active ligands
- PMID: 25609827
- PMCID: PMC4378623
- DOI: 10.1093/jxb/eru522
Plant membrane assays with cytokinin receptors underpin the unique role of free cytokinin bases as biologically active ligands
Abstract
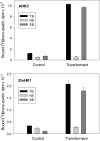
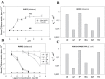
Cytokinin receptors play a key role in cytokinin-dependent processes regulating plant growth, development, and adaptation; therefore, the functional properties of these receptors are of great importance. Previously the properties of cytokinin receptors were investigated in heterologous assay systems using unicellular microorganisms, mainly bacteria, expressing receptor proteins. However, within microorganisms receptors reside in an alien environment that might distort the receptor properties. Therefore, a new assay system has been developed allowing studies of individual receptors within plant membranes (i.e. closer to their natural environment). The main ligand-binding characteristics of receptors from Arabidopsis [AHK2, AHK3, and AHK4] and maize (ZmHK1) were refined in this new system, and the properties of full-length Arabidopsis receptor AHK2 were characterized for the first time. Ligand specificity profiles of receptors towards cytokinin bases were comparable with the profiles retrieved in bacterial assay systems. In contrast, cytokinin-9-ribosides displayed a strongly reduced affinity for receptors in the plant assay system, indicating that ribosides as the common transport form of cytokinins have no or very weak cytokinin activity. This underpins the central role of free bases as the sole biologically active cytokinin compounds. According to molecular modelling and docking studies, N (9)-ribosylation alters the bonding pattern in cytokinin-receptor interaction and prevents β6-β7 loop movement important for tight hormone binding. A common feature of all receptors was a greatly reduced ligand binding at low (5.0-5.5) pH. The particularly high sensitivity of ZmHK1 to pH changes leads to the suggestion that some cytokinin receptors may play an additional role as pH sensors in the lumen of the endoplasmic reticulum.
Keywords: Arabidopsis thaliana; Zea mays.; cytokinin; cytokinin receptor; ligand specificity; pH sensor; plant assay system; receptor binding assay.
© The Author 2015. Published by Oxford University Press on behalf of the Society for Experimental Biology.
Figures




References
-
- Alvarez S, Marsh EL, Schroeder SG, Schachtman DP. 2008. Metabolomic and proteomic changes in the xylem sap of maize under drought. Plant, Cell and Environment 31, 325–340. - PubMed
-
- Bibikova TN, Jacob T, Dahse I, Gilroy S. 1998. Localized changes in apoplastic and cytoplasmic pH are associated with root hair development in Arabidopsis thaliana . Development 125, 2925–2934. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

