Complex regulation of γ-secretase: from obligatory to modulatory subunits
- PMID: 25610395
- PMCID: PMC4285130
- DOI: 10.3389/fnagi.2014.00342
Complex regulation of γ-secretase: from obligatory to modulatory subunits
Abstract
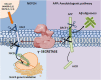
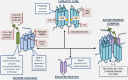
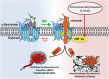
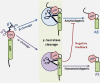
γ-Secretase is a four subunit, 19-pass transmembrane enzyme that cleaves amyloid precursor protein (APP), catalyzing the formation of amyloid beta (Aβ) peptides that form amyloid plaques, which contribute to Alzheimer's disease (AD) pathogenesis. γ-Secretase also cleaves Notch, among many other type I transmembrane substrates. Despite its seemingly promiscuous enzymatic capacity, γ-secretase activity is tightly regulated. This regulation is a function of many cellular entities, including but not limited to the essential γ-secretase subunits, nonessential (modulatory) subunits, and γ-secretase substrates. Regulation is also accomplished by an array of cellular events, such as presenilin (active subunit of γ-secretase) endoproteolysis and hypoxia. In this review we discuss how γ-secretase is regulated with the hope that an advanced understanding of these mechanisms will aid in the development of effective therapeutics for γ-secretase-associated diseases like AD and Notch-addicted cancer.
Keywords: APP; Alzheimer’s disease; Hif-1α; Notch; presenilin; β-amyloid; γ-Secretase.
Figures
References
-
- Alzforum (2014). GSAP revisited: does it really play a role in processing Aβ? http://www.alzforum.org/news/research-news/gsap-revisited-does-it-really...
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

