Distribution and frequencies of post-transcriptional modifications in tRNAs
- PMID: 25611331
- PMCID: PMC4615829
- DOI: 10.4161/15476286.2014.992273
Distribution and frequencies of post-transcriptional modifications in tRNAs
Abstract
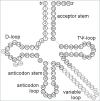
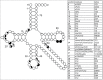
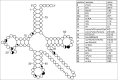
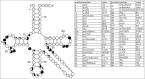
Functional tRNA molecules always contain a wide variety of post-transcriptionally modified nucleosides. These modifications stabilize tRNA structure, allow for proper interaction with other macromolecules and fine-tune the decoding of mRNAs during translation. Their presence in functionally important regions of tRNA is conserved in all domains of life. However, the identities of many of these modified residues depend much on the phylogeny of organisms the tRNAs are found in, attesting for domain-specific strategies of tRNA maturation. In this work we present a new tool, tRNAmodviz web server (http://genesilico.pl/trnamodviz) for easy comparative analysis and visualization of modification patterns in individual tRNAs, as well as in groups of selected tRNA sequences. We also present results of comparative analysis of tRNA sequences derived from 7 phylogenetically distinct groups of organisms: Gram-negative bacteria, Gram-positive bacteria, cytosol of eukaryotic single cell organisms, Fungi and Metazoa, cytosol of Viridiplantae, mitochondria, plastids and Euryarchaeota. These data update the study conducted 20 y ago with the tRNA sequences available at that time.
Keywords: RNA maturation; comparative analysis; evolution; modified nucleotides; post-transcriptional modification; tRNA; tRNA modifications; tRNA sequence; web server.
Figures


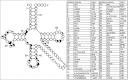
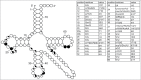
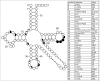
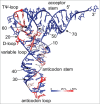
References
-
- Motorin Y, Helm M. tRNA stabilization by modified nucleotides. Biochemistry 2010; 49:4934-44; PMID:20459084; http://dx.doi.org/ 10.1021/bi100408z - DOI - PubMed
-
- Giegé R, Lapointe J. Transfer RNA aminoacylation and modified nucleosides. In: Grosjean H, ed. DNA and RNA Modification Enzymes: Structure, Mechanism, Function and Evolution. Austin, Texas, USA: Landes Bioscience, 2009:476-92
-
- Agris PF. Bringing order to translation: the contributions of transfer RNA anticodon-domain modifications. EMBO Rep 2008; 9:629-35; PMID:18552770; http://dx.doi.org/ 10.1038/embor.2008.104 - DOI - PMC - PubMed
-
- Grosjean H, de Crecy-Lagard V, Marck C. Deciphering synonymous codons in the three domains of life: co-evolution with specific tRNA modification enzymes. FEBS letters 2010; 584:252-64; PMID:19931533; http://dx.doi.org/ 10.1016/j.febslet.2009.11.052 - DOI - PubMed
-
- Helm M, Alfonzo JD. Posttranscriptional RNA Modifications: playing metabolic games in a cell's chemical Legoland. Chem Biol 2014; 21:174-85; PMID:24315934; http://dx.doi.org/ 10.1016/j.chembiol.2013.10.015 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
