NF-κB-dependent and -independent epigenetic modulation using the novel anti-cancer agent DMAPT
- PMID: 25611383
- PMCID: PMC4669767
- DOI: 10.1038/cddis.2014.569
NF-κB-dependent and -independent epigenetic modulation using the novel anti-cancer agent DMAPT
Abstract
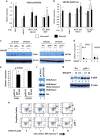
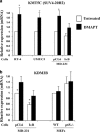
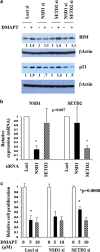
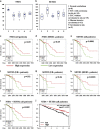
The transcription factor nuclear factor-kappaB (NF-κB) is constitutively active in several cancers and is a target of therapeutic development. We recently developed dimethylaminoparthenolide (DMAPT), a clinical grade water-soluble analog of parthenolide, as a potent inhibitor of NF-κB and demonstrated in vitro and in vivo anti-tumor activities in multiple cancers. In this study, we show DMAPT is an epigenetic modulator functioning in an NF-κB-dependent and -independent manner. DMAPT-mediated NF-κB inhibition resulted in elevated histone H3K36 trimethylation (H3K36me3), which could be recapitulated through genetic ablation of the p65 subunit of NF-κB or inhibitor-of-kappaB alpha super-repressor overexpression. DMAPT treatment and p65 ablation increased the levels of H3K36 trimethylases NSD1 (KMT3B) and SETD2 (KMT3A), suggesting that NF-κB directly represses their expression and that lower H3K36me3 is an epigenetic marker of constitutive NF-κB activity. Overexpression of a constitutively active p65 subunit of NF-κB reduced NSD1 and H3K36me3 levels. NSD1 is essential for DMAPT-induced expression of pro-apoptotic BIM, indicating a functional link between epigenetic modification and gene expression. Interestingly, we observed enhanced H4K20 trimethylation and induction of H4K20 trimethylase KMT5C in DMAPT-treated cells independent of NF-κB inhibition. These results add KMT5C to the list NF-κB-independent epigenetic targets of parthenolide, which include previously described histone deacetylase 1 (HDAC-1) and DNA methyltransferase 1. As NSD1 and SETD2 are known tumor suppressors and loss of H4K20 trimethylation is an early event in cancer progression, which contributes to genomic instability, we propose DMAPT as a potent pharmacologic agent that can reverse NF-κB-dependent and -independent cancer-specific epigenetic abnormalities.
Figures


References
-
- Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell 2007; 128: 735–745. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

