Genome-wide identification and characterization of the bHLH gene family in tomato
- PMID: 25612924
- PMCID: PMC4312455
- DOI: 10.1186/s12864-014-1209-2
Genome-wide identification and characterization of the bHLH gene family in tomato
Abstract
Background: The basic helix-loop-helix (bHLH) proteins are a large superfamily of transcription factors, and play a central role in a wide range of metabolic, physiological, and developmental processes in higher organisms. Tomato is an important vegetable crop, and its genome sequence has been published recently. However, the bHLH gene family of tomato has not been systematically identified and characterized yet.
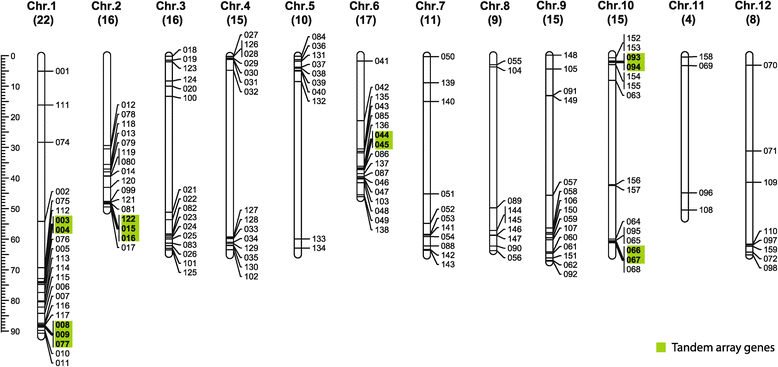
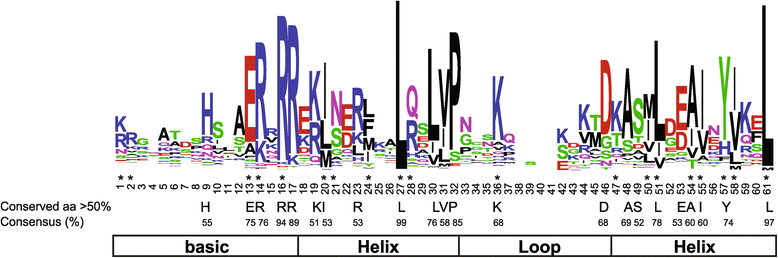
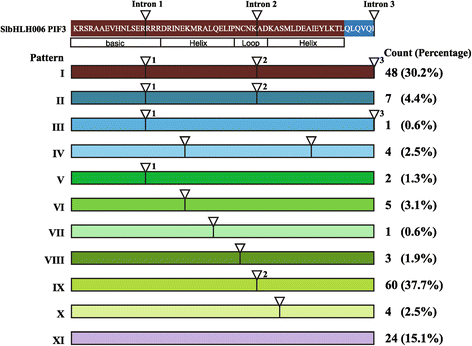
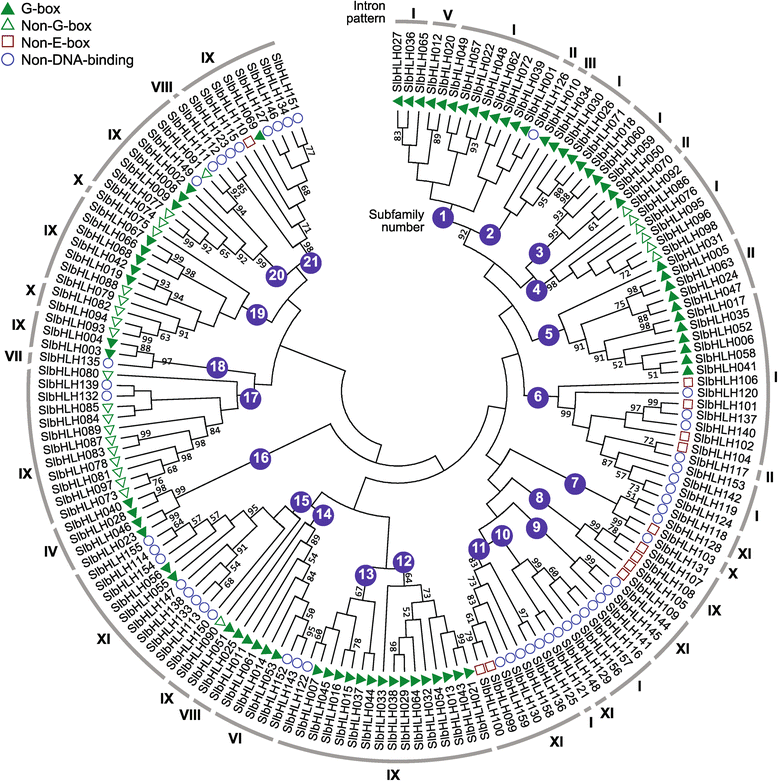
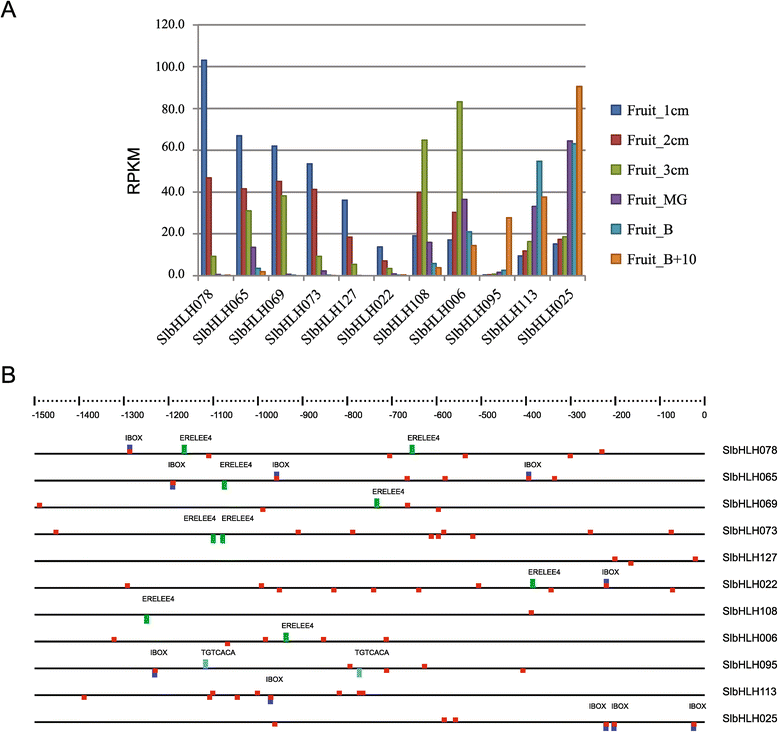
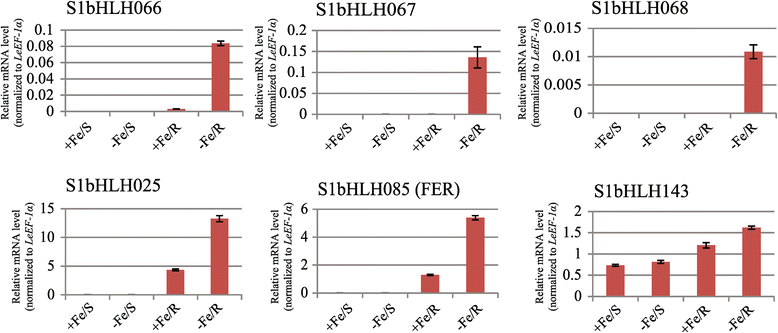
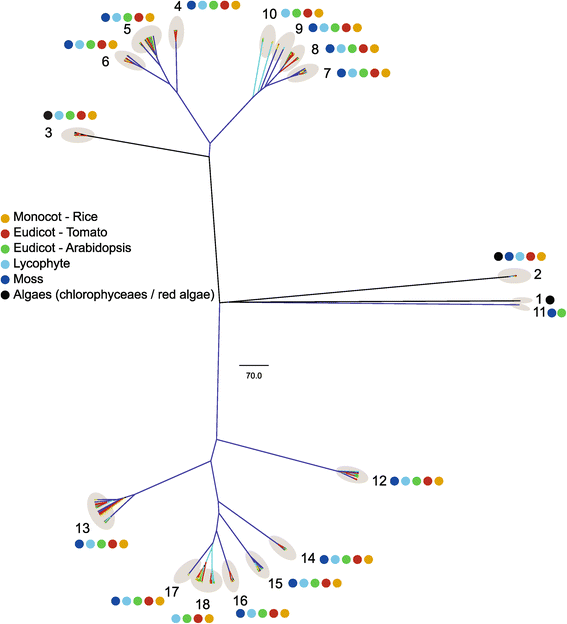
Results: In this study, we identified 159 bHLH protein-encoding genes (SlbHLH) in tomato genome and analyzed their structures. Although bHLH domains were conserved among the bHLH proteins between tomato and Arabidopsis, the intron sequences and distribution of tomato bHLH genes were extremely different compared with Arabidopsis. The gene duplication analysis showed that 58.5% and 6.3% of SlbHLH genes belonged to low-stringency and high-stringency duplication, respectively, indicating that the SlbHLH genes are mainly generated via short low-stringency region duplication in tomato. Subsequently, we classified the SlbHLH genes into 21 subfamilies by phylogenetic tree analysis, and predicted their possible functions by comparison with their homologous genes of Arabidopsis. Moreover, the expression profile analysis of SlbHLH genes from 10 different tissues showed that 21 SlbHLH genes exhibited tissue-specific expression. Further, we identified that 11 SlbHLH genes were associated with fruit development and ripening (eight of them associated with young fruit development and three with fruit ripening). The evolutionary analysis revealed that 92% SlbHLH genes might be evolved from ancestor(s) originated from early land plant, and 8% from algae.
Conclusions: In this work, we systematically identified SlbHLHs by analyzing the tomato genome sequence using a set of bioinformatics approaches, and characterized their chromosomal distribution, gene structures, duplication, phylogenetic relationship and expression profiles, as well predicted their possible biological functions via comparative analysis with bHLHs of Arabidopsis. The results and information provide a good basis for further investigation of the biological functions and evolution of tomato bHLH genes.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

