Muscle cell fate choice requires the T-box transcription factor midline in Drosophila
- PMID: 25614583
- PMCID: PMC4349071
- DOI: 10.1534/genetics.115.174300
Muscle cell fate choice requires the T-box transcription factor midline in Drosophila
Abstract
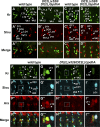
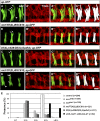
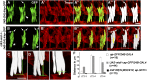
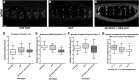
Drosophila Midline (Mid) is an ortholog of vertebrate Tbx20, which plays roles in the developing heart, migrating cranial motor neurons, and endothelial cells. Mid functions in cell-fate specification and differentiation of tissues that include the ectoderm, cardioblasts, neuroblasts, and egg chambers; however, a role in the somatic musculature has not been described. We identified mid in genetic and molecular screens for factors contributing to somatic muscle morphogenesis. Mid is expressed in founder cells (FCs) for several muscle fibers, and functions cooperatively with the T-box protein H15 in lateral oblique muscle 1 and the segment border muscle. Mid is particularly important for the specification and development of the lateral transverse (LT) muscles LT3 and LT4, which arise by asymmetric division of a single muscle progenitor. Mid is expressed in this progenitor and its two sibling FCs, but is maintained only in the LT4 FC. Both muscles were frequently missing in mid mutant embryos, and LT4-associated expression of the transcription factor Krüppel (Kr) was lost. When present, LT4 adopted an LT3-like morphology. Coordinately, mid misexpression caused LT3 to adopt an LT4-like morphology and was associated with ectopic Kr expression. From these data, we concluded that mid functions first in the progenitor to direct development of LT3 and LT4, and later in the FCs to influence whichever of these differentiation profiles is selected. Mid is the first T-box factor shown to influence LT3 and LT4 muscle identity and, along with the T-box protein Optomotor-blind-related-gene 1 (Org-1), is representative of a new class of transcription factors in muscle specification.
Keywords: Tbx20; differentiation; founder cell; midline; muscle identity.
Copyright © 2015 by the Genetics Society of America.
Figures



References
-
- Abmayr S. M., Keller C. A., 1998. Drosophila myogenesis and insights into the role of nautilus. Curr. Top. Dev. Biol. 38: 35–80. - PubMed
-
- Bate M., 1990. The embryonic development of larval muscles in Drosophila. Development 110: 791–804. - PubMed
-
- Baylies M. K., Bate M., 1996. twist: a myogenic switch in Drosophila. Science 272: 1481–1484. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R35 GM118068/GM/NIGMS NIH HHS/United States
- R01-AR44274/AR/NIAMS NIH HHS/United States
- F32 AR057290/AR/NIAMS NIH HHS/United States
- R01 AR044274/AR/NIAMS NIH HHS/United States
- R37 GM047867/GM/NIGMS NIH HHS/United States
- R01 AR068128/AR/NIAMS NIH HHS/United States
- R01 GM099945/GM/NIGMS NIH HHS/United States
- P30 CA008748/CA/NCI NIH HHS/United States
- 1F32AR057290/AR/NIAMS NIH HHS/United States
- R01 GM047867/GM/NIGMS NIH HHS/United States
- R01-GM56989/GM/NIGMS NIH HHS/United States
- R01 GM056989/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

