Escherichia coli and Staphylococcus phages: effect of translation initiation efficiency on differential codon adaptation mediated by virulent and temperate lifestyles
- PMID: 25614589
- PMCID: PMC4631060
- DOI: 10.1099/vir.0.000050
Escherichia coli and Staphylococcus phages: effect of translation initiation efficiency on differential codon adaptation mediated by virulent and temperate lifestyles
Abstract
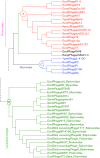
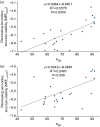
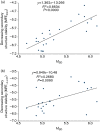
Rapid biosynthesis is key to the success of bacteria and viruses. Highly expressed genes in bacteria exhibit a strong codon bias corresponding to the differential availability of tRNAs. However, a large clade of lambdoid coliphages exhibits relatively poor codon adaptation to the host translation machinery, in contrast to other coliphages that exhibit strong codon adaptation to the host. Three possible explanations were previously proposed but dismissed: (1) the phage-borne tRNA genes that reduce the dependence of phage translation on host tRNAs, (2) lack of time needed for evolving codon adaptation due to recent host switching, and (3) strong strand asymmetry with biased mutation disrupting codon adaptation. Here, we examined the possibility that phages with relatively poor codon adaptation have poor translation initiation which would weaken the selection on codon adaptation. We measured translation initiation by: (1) the strength and position of the Shine-Dalgarno (SD) sequence, and (2) the stability of the secondary structure of sequences flanking the SD and start codon known to affect accessibility of the SD sequence and start codon. Phage genes with strong codon adaptation had significantly stronger SD sequences than those with poor codon adaptation. The former also had significantly weaker secondary structure in sequences flanking the SD sequence and start codon than the latter. Thus, lambdoid phages do not exhibit strong codon adaptation because they have relatively inefficient translation initiation and would benefit little from increased elongation efficiency. We also provided evidence suggesting that phage lifestyle (virulent versus temperate) affected selection intensity on the efficiency of translation initiation and elongation.
© 2015 The Authors.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

