Genetic characterization of human coxsackievirus A6 variants associated with atypical hand, foot and mouth disease: a potential role of recombination in emergence and pathogenicity
- PMID: 25614593
- PMCID: PMC4631059
- DOI: 10.1099/vir.0.000062
Genetic characterization of human coxsackievirus A6 variants associated with atypical hand, foot and mouth disease: a potential role of recombination in emergence and pathogenicity
Abstract
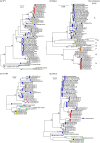
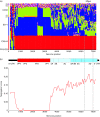
Human coxsackievirus A6 (CVA6) is an enterically transmitted enterovirus. Until recently, CVA6 infections were considered as being of minor clinical significance, and only rarely aetiologically linked with hand, foot and mouth disease (HFMD) associated with other species A enteroviruses (particularly EV71 and CVA16). From 2008 onwards, however, CVA6 infections have been associated with several outbreaks worldwide of atypical HFMD (aHFMD) accompanied by a varicelliform rash. We recently reported CVA6-associated eczema herpeticum occurring predominantly in children and young adults in Edinburgh in January and February 2014. To investigate genetic determinants of novel clinical phenotypes of CVA6, we genetically characterized and analysed CVA6 variants associated with eczema herpeticum in Edinburgh in 2014 and those with aHFMD in CAV isolates collected from 2008. A total of eight recombinant forms (RFs) have circulated worldwide over the past 10 years, with the particularly recent appearance of RF-H associated with eczema herpeticum cases in Edinburgh in 2014. Comparison of phylogenies and divergence of complete genome sequences of CVA6 identified recombination breakpoints in 2A-2C, within VP3, and between 5' untranslated region and VP1. A Bayesian temporal reconstruction of CVA6 evolution since 2004 provided estimates of dates and the actual recombination events that generated more recently appearing recombination groups (RF-E, -F, -G and -H). Associations were observed between recombination groups and clinical presentations of herpangina, aHFMD and eczema herpeticum, but not with VP1 or other structural genes. These observations provided evidence that NS gene regions may potentially contribute to clinical phenotypes and outcomes of CVA6 infection.
© 2015 The Authors.
Figures




References
-
- Bailly J. L., Mirand A., Henquell C., Archimbaud C., Chambon M., Charbonné F., Traoré O., Peigue-Lafeuille H. ( 2009). Phylogeography of circulating populations of human echovirus 30 over 50 years: nucleotide polymorphism and signature of purifying selection in the VP1 capsid protein gene. Infect Genet Evol 9, 699–708. 10.1016/j.meegid.2008.04.009 - DOI - PubMed
-
- Chung W. H., Shih S. R., Chang C. F., Lin T. Y., Huang Y. C., Chang S. C., Liu M. T., Ko Y. S., Deng M. C., et al. ( 2013). Clinicopathologic analysis of coxsackievirus A6 new variant induced widespread mucocutaneous bullous reactions mimicking severe cutaneous adverse reactions. J Infect Dis 208, 1968–1978. 10.1093/infdis/jit383 - DOI - PubMed
-
- Costa-Mattioli M., Ferré V., Casane D., Perez-Bercoff R., Coste-Burel M., Imbert-Marcille B. M., Andre E. C., Bressollette-Bodin C., Billaudel S., Cristina J. ( 2003). Evidence of recombination in natural populations of hepatitis A virus. Virology 311, 51–59. 10.1016/S0042-6822(03)00109-0 - DOI - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

